Spatial and Temporal Scales Matter When Assessing the Species and Genetic Diversity of Springtails (Collembola) in Antarctica
- 1School of Science, University of Waikato, Hamilton, New Zealand
- 2Polar Knowledge Canada, Canadian High Arctic Research Station, Cambridge Bay, NU, Canada
- 3British Antarctic Survey, Cambridge, United Kingdom
Seven species of springtail (Collembola) are present in Victoria Land, Antarctica and all have now been sequenced at the DNA barcoding region of the mitochondrial cytochrome c oxidase subunit I gene (COI). Here, we review these sequence data (n = 930) from the GenBank and Barcode of Life Datasystems (BOLD) online databases and provide additional, previously unpublished sequences (n = 392) to assess the geographic distribution of COI variants across all species. Four species (Kaylathalia klovstadi, Cryptopygus cisantarcticus, Friesea grisea, and Cryptopygus terranovus) are restricted to northern Victoria Land and three (Antarcticinella monoculata, Cryptopygus nivicolus, and Gomphiocephalus hodgsoni) are found only in southern Victoria Land, the two biogeographic zones which are separated by the vicinity of the Drygalski Ice Tongue. We found highly divergent lineages within all seven species (range 1.7–14.7%) corresponding to different geographic locations. Levels of genetic divergence for the southern Victoria Land species G. hodgsoni, the most widespread species (~27,000 km2), ranged from 5.9 to 7.3% divergence at sites located within 30 km, but separated by glaciers. We also found that the spatial patterns of genetic divergence differed between species. For example, levels of divergence were much higher for C. terranovus (>10%) than for F. grisea (<0.2%) that had been collected from the same sites in northern Victoria Land. Glaciers have been suggested to be major barriers to dispersal and two species (C. cisantarcticus and F. grisea) showed highly divergent (>5%) populations and over 87% of the total genetic variation (based on AMOVA) on either side of a single, 16 km width glacier. Collectively, these data provide evidence for limited dispersal opportunities among populations of springtails due to geological and glaciological barriers (e.g., glaciers and ice tongues). Some locations harbored highly genetically divergent populations and these areas are highlighted from a conservation perspective such as avoidance of human-mediated transport between sites. We conclude that species-specific spatial and temporal scales need to be considered when addressing ecological and physiological questions as well as conservation strategies for Antarctic Collembola.
Introduction
Due to the extreme environmental conditions that characterize Antarctica (Convey, 2013), as well as the geographical, and island-like, isolation of suitable terrestrial habitats (Bergstrom and Chown, 1999), long-range dispersal events for Antarctic Collembola (springtails) are rare and they usually rely on liquid water for transport via flotation (Hawes et al., 2008; McGaughran et al., 2011a; Carapelli et al., 2017). It is likely that springtails are currently unable to disperse among the three Antarctic Conservation Biogeographic Regions (ACBRs; Terauds and Lee, 2016) in the Ross Sea Sector of Antarctica: northern Victoria Land (NVL), southern Victoria Land (SVL), and the Transantarctic Mountains (TAM), owing to various physical obstacles in the marine and terrestrial realms (e.g., the Drygalski Ice Tongue; Figure 1). Within each of these three ACBRs, available habitat is patchy and local microhabitats are likely to be important for long-term persistence of populations (Sinclair and Sjursen, 2001). These small “island” populations can thus accumulate genetic mutations resulting in distinct genetic patterns across the landscape. Previous studies on the diversity patterns of lichen communities within the Antarctic have suggested that distributions are likely to have been driven by geological or glaciological features rather than environmental gradients that are associated with latitudinal or longitudinal distances (Adams et al., 2006; Peat et al., 2007; Green et al., 2011; Colesie et al., 2014). Similarly, the distributions of Antarctic springtail species may not follow a pattern of decreasing species diversity with increasing latitude (e.g., Caruso et al., 2009). The spatial scales at which sampling is undertaken are, therefore, important as genetic differences between populations do not necessarily increase proportionately with distance. However, until now a broader synthesis of genetic data for multiple species across larger spatial scales has not been undertaken. This is unfortunate as such an analysis could help to assess the relative role of geographic barriers in structuring populations of Antarctic springtails.
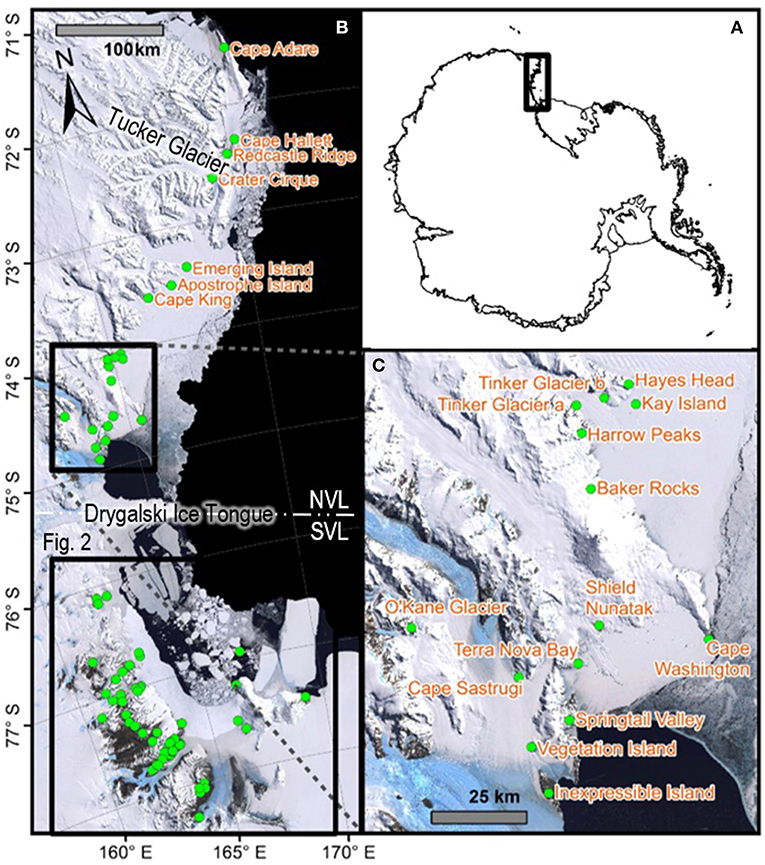
Figure 1. Map with all 21 sites in northern Victoria Land labeled, including (A) an overview map showing the location of Victoria Land within the Antarctic continent, (B) all 88 sites within Victoria Land from which COI sequences were obtained and presented in the current study, for the seven species in this area, and (C) a zoomed-in portion of NVL to display site names. Generated in ArcMap v10.5.1. See Table 2 and Table S1 for GPS co-ordinates.
Terrestrial invertebrates were first discovered in the Ross Sea sector of Antarctica during the early 1900s (e.g., Carpenter, 1902, 1908; Willem, 1902; Gregory, 1909; Macnamara, 1919). Further entomological research was undertaken during the 1960s and 1970s and provided morphological descriptions and distributional ranges as well as general ecological observations and physiological studies, particularly for the most widespread species in this area—Gomphiocephalus hodgsoni (Gressitt and Leech, 1961; Salmon, 1962; Gressitt et al., 1963; Janetschek, 1963, 1967, 1970). More recently, molecular approaches including allozyme analyses (e.g., Frati et al., 1996; Stevens and Hogg, 2003) and DNA sequencing of mitochondrial cytochrome c oxidase subunit II (e.g., Frati and Dell'Ampio, 2000; Frati et al., 2001; McGaughran et al., 2010) and cytochrome c oxidase subunit I (COI) gene regions (e.g., Nolan et al., 2006; Stevens and Hogg, 2006; McGaughran et al., 2010) have been undertaken for Antarctic springtails. These data have enhanced understanding of their evolutionary histories and allowed testing of hypotheses, such as endemism of the fauna, proposed by earlier researchers (e.g., Wise, 1967). Further, they have provided insights into the long-term persistence of endemic taxa, their survival in refugia and subsequent expansion during interglacial periods (e.g., Convey et al., 2008; McGaughran et al., 2011b; Beet et al., 2016; Carapelli et al., 2017). In turn, this has facilitated predictions as to how populations may respond to future changes in habitat availability and environmental conditions (e.g., Chown and Convey, 2007; Lee et al., 2017). While these studies either focus on a single species or have made comparisons between two species, none has assessed the full range of COI sequences available for all known species in the area.
Based on early studies of springtail distribution using morphological taxonomy, the Ross Sea sector (longitude 160–175°E) was subdivided into five areas (Wise, 1967), although more recent data have confirmed that some of the described species distributions within these five areas were incorrect (e.g., Ross Island contains only G. hodgsoni). Studies focused on specific taxa (e.g., Cryptopygus terranovus) have suggested additional landscape divisions based on genetic evidence for geographical isolation (Stevens et al., 2006b; Hawes et al., 2010; Carapelli et al., 2017). However, other species present in the same area do not necessarily exhibit the same divergence patterns (Caruso et al., 2009). For example, in an area of NVL where Friesea grisea and C. terranovus have overlapping ranges, the two species have very different levels of genetic divergence; <0.2% for F. grisea (Torricelli et al., 2010b) and >10% for C. terranovus (Carapelli et al., 2017).
Our current study compiled and consolidated all available COI sequences and collection information for each of the seven species found in Victoria Land; Cryptopygus cisantarcticus, C. terranovus, Friesea grisea, and Kaylathalia klovstadi in NVL, and Antarcticinella monoculata, C. nivicolus, and Gomphiocephalus hodgsoni in SVL (Table 1). We provide a comprehensive list of collection sites for each haplotype to assess spatial variability. We also examined whether spatially isolated populations would be distinct at the COI gene region, and if so, whether landscape features (such as particularly large glaciers and ice tongues) may provide contemporary barriers to dispersal. Where relevant, we highlight instances of possible cryptic speciation as indicated by highly distinct genetic lineages, as well as identify priority sites from a conservation perspective.
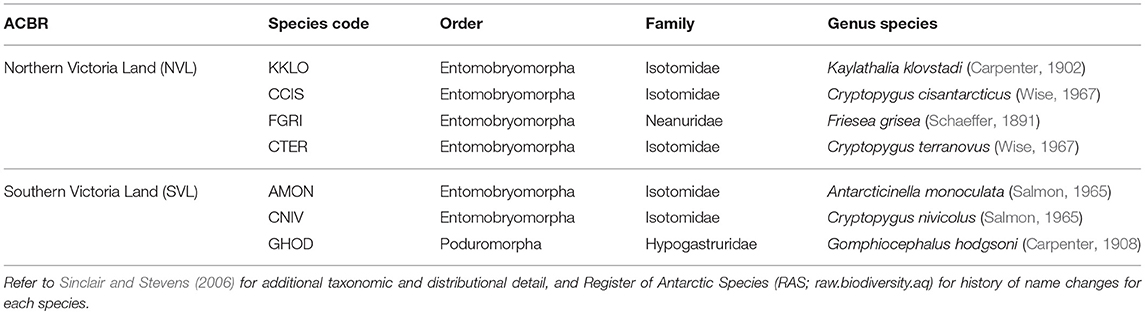
Table 1. The seven springtail species present in the two Antarctic Conservation Biogeographic Regions (ACBRs) in Victoria Land (NVL and SVL), within the Ross Sea sector of Antarctica.
Methods
Existing Sequence Data
All publicly available sequences were downloaded from the BOLD and GenBank databases (n = 930 sequences), covering the seven springtail species present in Victoria Land. Supporting information was also downloaded from BOLD where possible, although in many instances these data were absent, particularly for sequences that had been obtained directly from GenBank. Where possible, we endeavored to manually retrieve collection details from original research articles. To maximize sequence length of previously-analyzed specimens, original trace files (where possible) were imported into Geneious v11.1.5 (https://www.geneious.com) for complete re-analysis. Forward and reverse sequence reads were assembled, any discrepancies were edited and then consensus sequences were extracted. For cases where no trace files were available, final sequences that had been uploaded to GenBank or BOLD were used.
New Sequence Data
A total of 392 previously unpublished sequences have been included in the current study (Tables 2–4 and Table S2). Of these, 56 were obtained from individuals collected from NVL in January 2004 (C. Beard and R. Seppelt), January 2015 (C. Cary) and November 2017 (I. Hogg). For previously unpublished sequences of G. hodgsoni, specimens were collected from sites in the vicinities of Mackay Glacier (n = 60), Taylor Valley (n = 21) and the southern Dry Valleys (n = 249) from 2009 to 2016 (I. Hogg, G. Collins, C. Beet and N. Demetras). An additional six specimens of C. nivicolus were collected from Mount Seuss in 2008 (I. Hogg), and Mount Seuss and Tiger Island in 2015 (G. Collins, C. Beet, I. Hogg and D. Cowan). For a full list of sites where specimens were collected, see Figures 1, 2; Tables 2, 3, and Tables S1–S9.
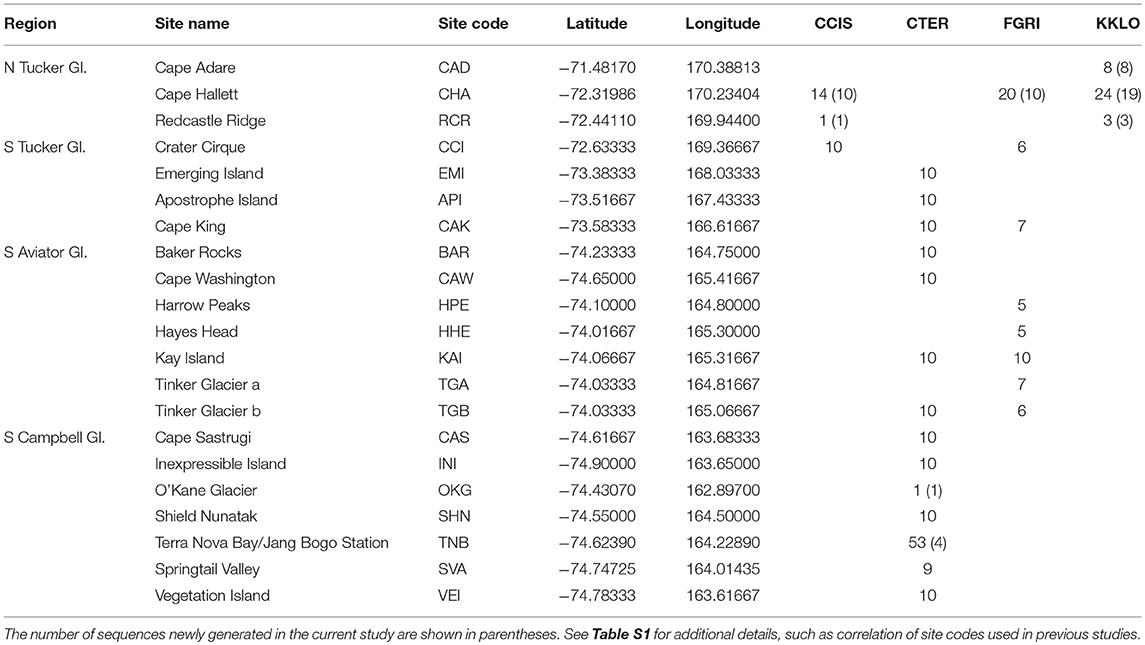
Table 2. Collection sites in northern Victoria Land, grouped by region, including the number of COI sequences at each site for the four species in this area; Cryptopygus cisantarcticus (CCIS), Cryptopygus terranovus (CTER), Friesea grisea (FGRI), and Kaylathalia klovstadi (KKLO).
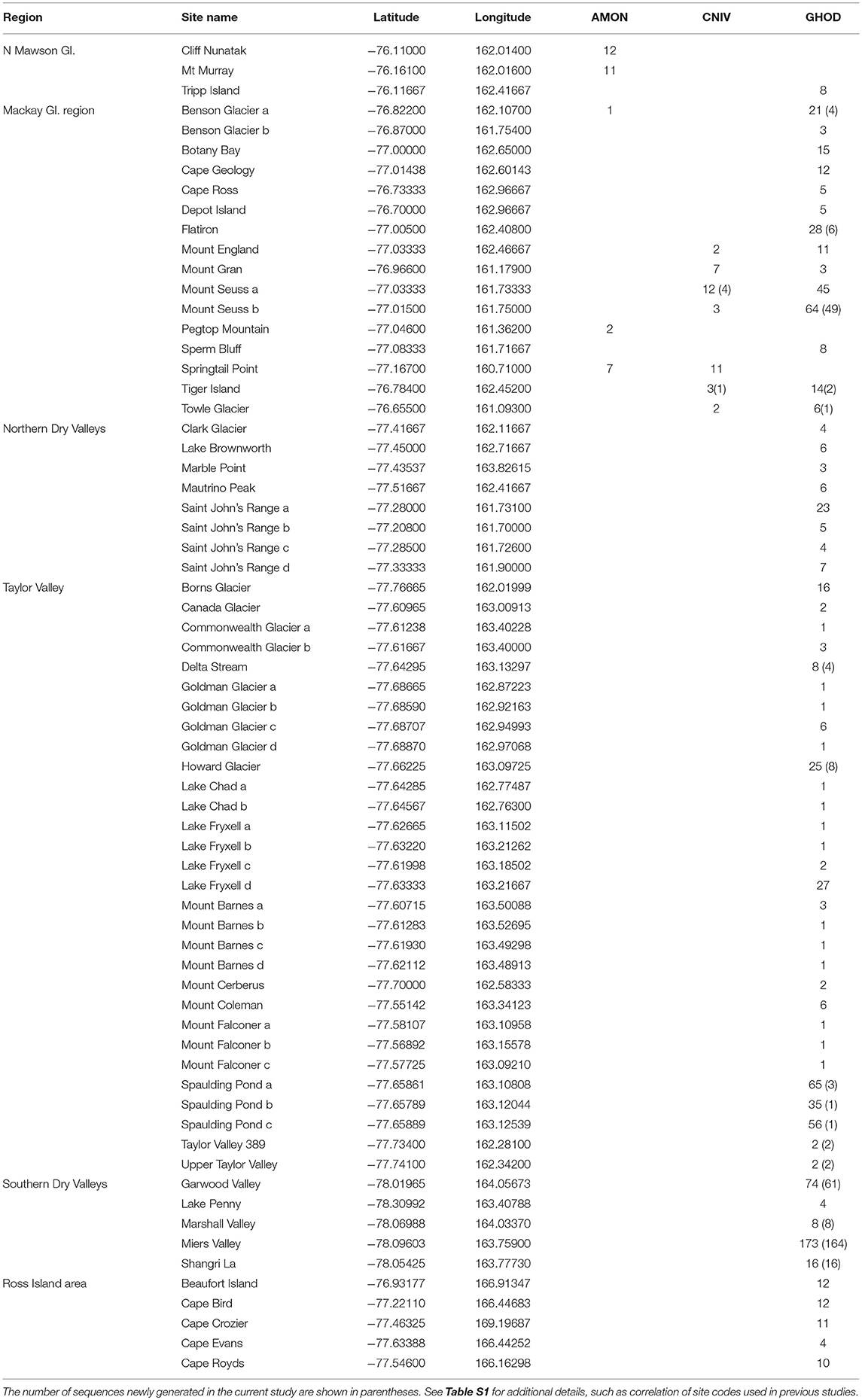
Table 3. List of collection sites in southern Victoria Land, grouped by region, including the number of COI sequences at each site for the three species in this area; Antarcticinella monoculata (AMON), Cryptopygus nivicolus (CNIV) and Gomphiocephalus hodgsoni (GHOD).
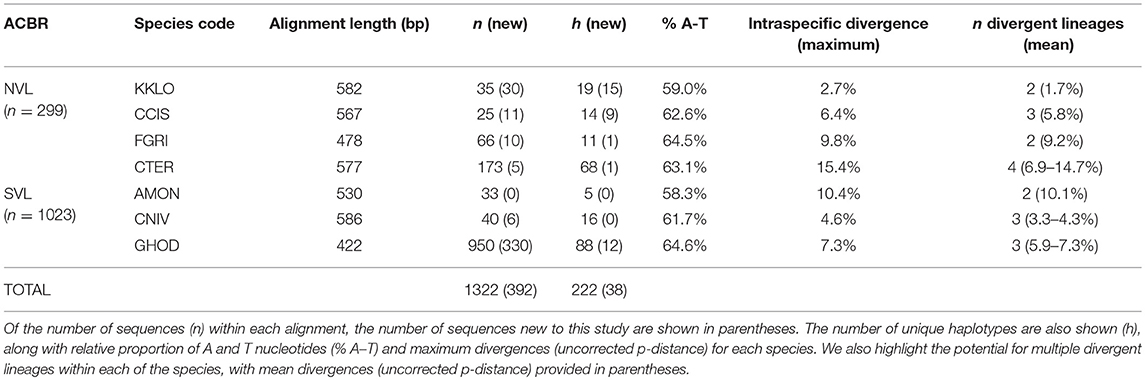
Table 4. Sequence details for the seven species of Collembola present in NVL and SVL, showing alignment lengths used in the current study (which vary for each species).
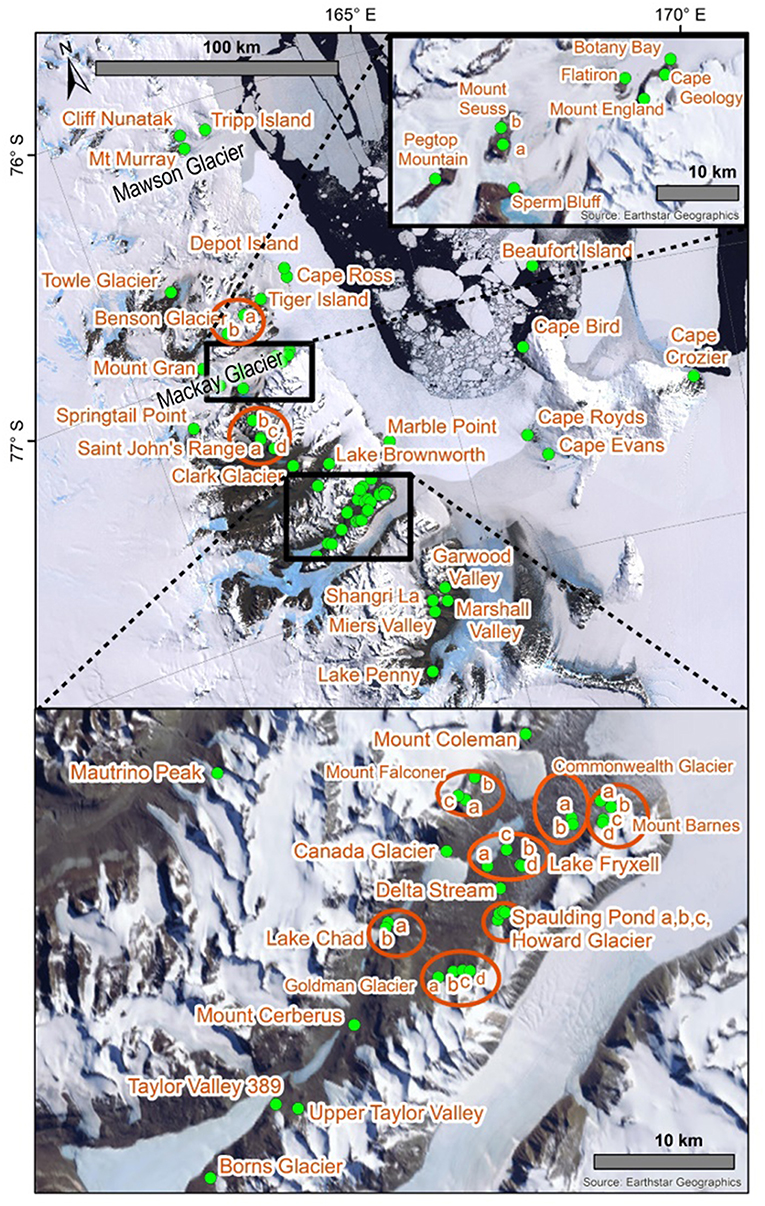
Figure 2. Map with all 67 sites in southern Victoria Land labeled, generated in ArcMap v10.5.1. See Table 3 and Table S1 for GPS co-ordinates.
These previously unpublished sequences were from specimens collected in pitfall traps (described in McGaughran et al., 2011a), using modified aspirators (Stevens and Hogg, 2002), or isolated by flotation from soil samples (Freckman and Virginia, 1993). In all cases, individual specimens were immediately preserved in 100% ethanol for later DNA extraction. DNA extractions, PCR amplifications and COI sequencing were carried out at the University of Waikato following procedures outlined in Collins and Hogg (2015) or at the Canadian Centre for DNA Barcoding following established protocols (see http://ccdb.ca/resources/).
Data Analyses
A separate alignment was made for each species using ClustalW within Geneious. After forward (LCO or LepF1) and reverse (HCO or LepR1) primer regions (26 bp each) were removed, each species' alignment was individually assessed and trimmed accordingly to maximize alignment length but also retain maximum sequence coverage for each species. Final alignments varied in length, from 422 to 586 bp. Based on these final trimmed alignments, unique haplotypes for each species were then manually assigned codes in order of the date they were sequenced (e.g., GHOD-001 for Gomphiocephalus hodgsoni, haplotype 1). Future haplotypes can therefore be added sequentially to this dataset.
The numbers of duplicate sequences for each haplotype were manually calculated based on the number of replicates within our trimmed alignments, as well as from original tables and figures provided in published papers for cases where only unique sequences were originally deposited online (Nolan et al., 2006; Demetras, 2010; McGaughran et al., 2010). This was particularly problematic for G. hodgsoni, as the numbers of each haplotype from each specific location could only be determined by a process of elimination through cross-referencing all available tables, figures and supplementary data. Unfortunately, collection information is absent for 46 G. hodgsoni specimens (see Table S9), outlined as follows. The precise collection location is uncertain for the two full mitochondrial genomes that have been previously assembled for G. hodgsoni (GenBank accessions AY191995 and NC005438; Nardi et al., 2003) as well as the three sequences from Greenslade et al. (2011). GenBank accession numbers for the five sequences new to McGaughran et al. (2008) could not be linked with their respective site data, and 41 specimens from McGaughran et al. (2010) which include those from McGaughran et al. (2008) were also lacking site data. For specimens from Demetras (2010) where collection data were not resolved to precise site, we have used the broader “valley-level” locations (i.e., Garwood, Marshall and Miers Valleys, and Shangri La) in our analyses. Overall, sequences from a total of 88 sites were assessed in this study (Figures 1, 2; Tables 2, 3 and Table S1). For specimens with collection data absent, sequences were included in the initial alignment for phylogenetic tree construction and haplotype assignments, and were then removed for subsequent biogeographic analyses.
Each of the individual alignments for the seven species were then assembled into one master alignment which was trimmed to 422 bp and a Maximum Likelihood tree was generated in MEGA v7.0.26 (Kumar et al., 2016) based on the sequence model GTR+I+G (Guindon and Gascuel, 2003; Darriba et al., 2012; JModelTest2), including 1,000 bootstrap replicates. All sequence divergence values included in the current study were also generated in MEGA based on uncorrected p-distances.
Haplotype pie charts (Figures 4–7) were generated in R v3.5.1 utilizing the packages “mapdata” (Becker et al., 2016) and “mapplots” (Gerritsen, 2014) based on the individual species alignments which were each trimmed to different lengths, depending on the sequences. To retain haplotype-level resolution, phylogenetic tree excerpts included in these figures were based on the individual species alignments (422–586 bp).
To determine potential barriers to dispersal, Analysis of Molecular Variance (AMOVA) analyses were performed separately for each species in R v3.5.1 utilizing the package “poppr” with 16,000 permutations (Kamvar et al., 2014, 2015). For this, species alignments that had each been trimmed to between 422 and 586 bp (Table 4) were used, and haplotypes were grouped according to their occurrence at sites within particular regions (listed in Tables 2, 3) to test the amount of genetic variability occurring within and between regions. As specimens of Cryptopygus nivicolus were all found within the Mackay Glacier region, we performed the AMOVA for this species with the sites Towle Glacier and Springtail Point designated as separate regions.
Results and Discussion
We compiled all available COI sequences for the seven springtail species in Victoria Land. Sequence coverage ranged from 25 to 950 individuals per species (total n = 1,322), with between 5 and 88 haplotypes identified for each species (Figure 3; Table 4). The final alignments for each species varied in length (422–586 bp), largely due to variation in sequence quality, and we treated each species separately for biogeographic analyses to maximize variability within the datasets, although a final alignment trimmed to 422 bp was used to construct the phylogenetic tree (Figure 3). Relative proportions of AT-richness ranged from 58.3 to 64.6% for each species (Table 4), and no insertions or deletions were found. Maximum intraspecific sequence divergences ranged from 2.7 to 15.4% (uncorrected p-distance) and for all seven species, high divergence values (1.7–14.7%) were found among individuals from different geographic locations (Table 4), supported by AMOVAs.
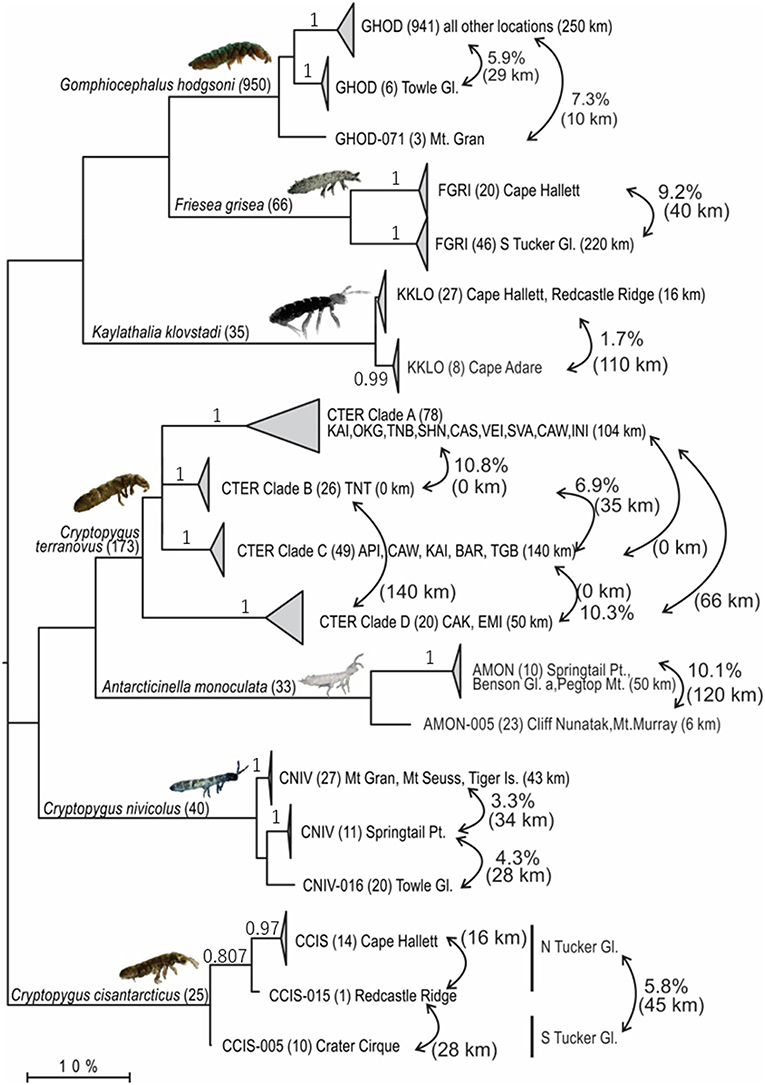
Figure 3. Maximum Likelihood tree (GTR + Γ + I; 1,000 bootstrap replicates) based on the trimmed alignment (422 bp) for all 1,322 COI sequences available for the seven springtail species in Victoria Land. Clades are collapsed proportionately, where each collapsed clade represents a potential cryptic species (>1.7% divergence), and number of sequences are shown in parentheses. Geographic distances within and between clades are provided, along with p-distances between clades, to highlight geographical isolation of well-supported clades (bootstrap values provided on branches, where relevant).
Northern Victoria Land Taxa
Northern Victoria Land is the northern-most Antarctic Conservation Biogeographic Region (ACBR) of the Ross Sea sector and extends from Cape Adare (71.3°S) to the Drygalski Ice Tongue (75.3°S) (Figure 1). We recovered a total of 299 COI sequences from the four springtail species that are present in this area; Kaylathalia klovstadi (KKLO; n = 35 sequences), Cryptopygus cisantarcticus (CCIS; n = 25), Friesea grisea (FGRI; n = 66), and Cryptopygus terranovus (CTER; n = 173). Each species was collected from a different range of sampling locations and also showed contrasting population genetic structures. For example, two genetically distinct lineages (clades C and D) of C. terranovus from sites in the central and northern zones as designated by previous studies (e.g., Fanciulli et al., 2001; Hawes et al., 2010; Carapelli et al., 2017), were highly divergent (>10%) whereas F. grisea specimens from the same area all had identical sequences (Figures 4, 5). Similarly, sequences of K. klovstadi were only 0.7% divergent between Cape Hallett and Redcastle Ridge, whereas C. cisantarcticus specimens differed by 4.3% between the same two collection sites. The Tucker Glacier is a significant barrier to dispersal for Antarctic springtails in the area, with highly distinct (>5%) populations present on either side of this glacier and no shared haplotypes between populations, consistent across species.
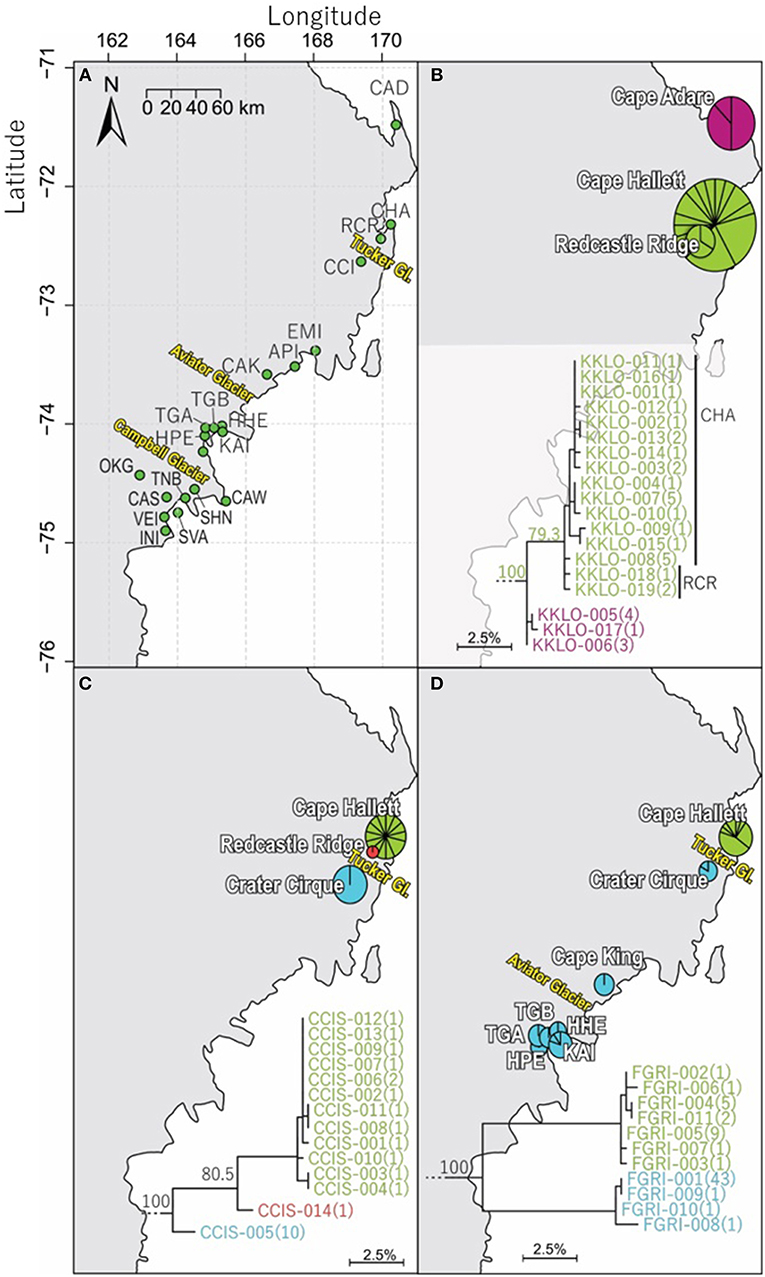
Figure 4. Geographical distribution of COI sequences for three of the four springtail species within northern Victoria Land, with (A) an overview of collection sites and major glacial barriers, (B) geographical distribution of the 19 haplotypes of Kaylathalia klovstadi (KKLO; n = 35; 582 bp), (C) geographical distribution of the 14 haplotypes of Cryptopygus cisantarcticus (CCIS; n = 25; 567 bp), and (D) geographical distribution of the 11 haplotypes of Friesea grisea (FGRI; n = 66; 478 bp). Pie charts are proportionate to the number of sequences, centered at collection site co-ordinates. Tucker Glacier is a major barrier of dispersal for CCIS and FGRI, and no sequences of KKLO have been obtained from south of the Tucker Glacier.
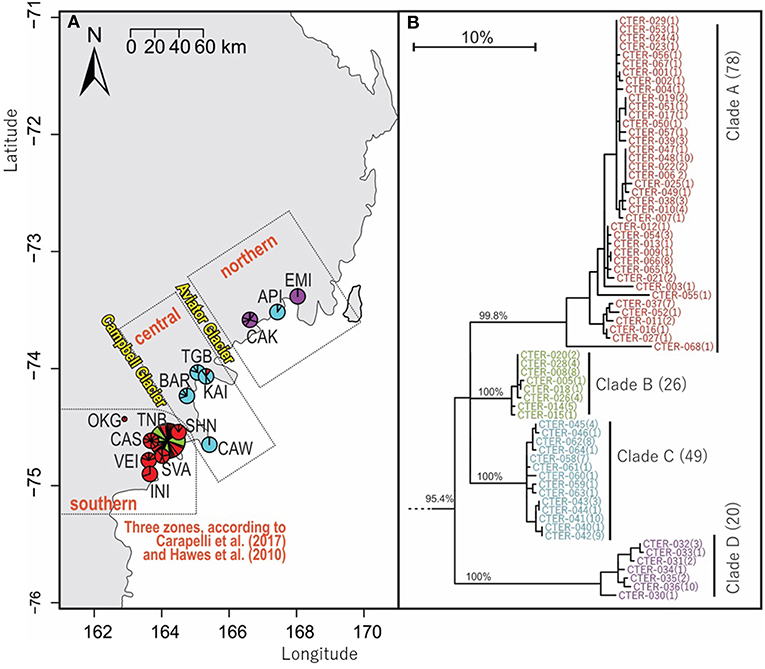
Figure 5. Geographical distribution within northern Victoria Land of COI sequences for Cryptopygus terranovus (CTER; n = 173; 577 bp) showing (A) distributional barriers (Campbell and Aviator Glaciers), and (B) clade groupings as previously suggested by Carapelli et al. (2017) and Hawes et al. (2010). Pie charts are proportionate to the number of sequences, centered at collection site co-ordinates.
Kaylathalia klovstadi (Carpenter, 1902)
The first collections of this species (originally described as Isotoma klovstadi) were obtained from Geikie Ridge during the British Antarctic Expedition (1898–1901), after which it was not collected again until 1965 from Ridley Beach at Cape Adare (Wise, 1971). The first five COI sequences obtained for K. klovstadi were from individuals collected at Cape Hallett and published in a study that reclassified the genus from Isotoma to Desoria (Stevens et al., 2006a). The same five sequences were further used to again reclassify the genus to Kaylathalia (Stevens and D'Haese, 2016).
We now include a further 30 previously unpublished COI sequences from three locations (including Cape Hallett), revealing 15 new haplotypes (KKLO-005 to KKLO-019; Tables 2, 4; Tables S2, S3). By expanding the sequence coverage and including two additional sites, we are now able to demonstrate geographical isolation of distinct genetic lineages (2%) for K. klovstadi (Figure 4B). When haplotypes in the combined dataset were grouped by region, an AMOVA showed that 75% of the sequence variation occurred between the northern Cape Adare group (n = 8), and the southern Cape Hallett and Redcastle Ridge group (n = 27) (Table 5; p < 0.01). No haplotypes were shared between sites, although Redcastle Ridge was represented by only 3 sequences which were similar to those from Cape Hallett (0.7% divergence).
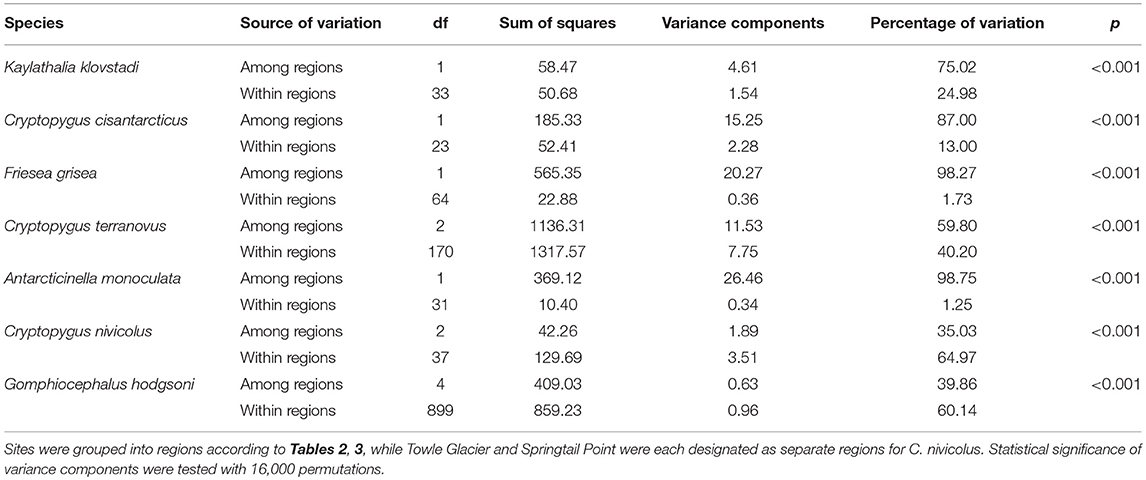
Table 5. Analysis of molecular variance (AMOVA) (Excoffier et al., 1992) results for the seven species of Collembola in Victoria Land, as implemented in R v3.5.1 using the package “poppr” (Kamvar et al., 2014, 2015).
Based on a study of 40 COII gene sequences, Frati et al. (2001) suggested that the Tucker Glacier was as a major barrier to dispersal for K. klovstadi. Currently, no COI sequences were available for K. klovstadi from the southern side of the Tucker Glacier and this would benefit from further attention, particularly as strong population genetic structure for the COI gene was observed for C. cisantarcticus and F. grisea on opposing sides of this glacier (see sections Cryptopygus cisantarcticus Wise, 1967 and Friesea grisea Schaeffer, 1891).
Cryptopygus cisantarcticus (Wise, 1967)
The first four COI sequences of C. cisantarcticus were from specimens collected at Cape Hallett and included in Stevens et al. (2006b), who suggested this species has been isolated for 18–11 MY based on comparisons with other Southern Hemisphere springtail species. An additional 10 sequences were obtained from Crater Cirque and presented as an outgroup in Carapelli et al. (2017).
Here, we added a further 10 sequences from Cape Hallett and a single sequence from Redcastle Ridge, representing nine new haplotypes (CCIS-006 to CCIS-014), and provide comparison among all COI sequences currently available for C. cisantarcticus (Tables 2, 4; Tables S2, S4). When haplotypes in the combined dataset were grouped by location, an AMOVA showed that 87% of the sequence variation occurred across the Tucker Glacier, between the northern Cape Hallett and Redcastle Ridge group (n = 15) and the southern Crater Cirque population (n = 10) (Table 5; p < 0.01).
Population genetic structure differed for C. cisantarcticus as compared to other species in the area where their sequence distributions overlapped (Figure 4). Sites at Redcastle Ridge and Cape Hallett are in relatively close proximity (<16 km), and divergence between these two locations was very low for K. klovstadi (0.7%) while much higher for C. cisantarcticus (4.3%). Furthermore, mean divergence for C. cisantarcticus across the Tucker Glacier was 5.8%, whereas divergence was higher for F. grisea, at >9%.
Friesea grisea (Schaeffer, 1891)
This species has been considered “pan-Antarctic” until very recently; specimens of Friesea grisea from the Antarctic Peninsula were found to be morphologically distinct to those in Eastern Antarctica (Greenslade, 2018) in agreement with the absence of haplotype sharing between the two locations (>15% divergence; Torricelli et al., 2010b). A total of 55 COI sequences for F. grisea have been published (Torricelli et al., 2010b). In addition, the full mitochondrial genome has previously been assembled for F. grisea (GenBank accession KR180288) collected from Kay Island (Torricelli et al., 2010a) and we have included the COI gene region from this sequence in our analyses.
We added an additional 10 unpublished sequences from Cape Hallett (situated to the north of Tucker Glacier; Figure 4), including one new haplotype, for a total of 20 sequences from this site (Table 2; Tables S2, S5). When haplotypes in the combined dataset were grouped by location, an AMOVA showed that 98.3% of the sequence variation occurred across the Tucker Glacier (Table 5; p < 0.01), between the northern Cape Hallett population (n = 20) and the southern group (n = 46) comprised of seven sites across approximately 220 km of coastal terrain (Figure 4D). Individuals of F. grisea along that entire coastal area south of the Tucker Glacier were within 0.2% divergence, while the population at Cape Hallett was highly differentiated (9.2% divergence), further supporting a lack of dispersal across the Tucker Glacier, in agreement with our findings for C. cisantarcticus.
Cryptopygus terranovus (Wise, 1967)
An analysis of allozymes for Cryptopygus terranovus (previously known as Gressittacantha terranova; Greenslade, 2015) showed northern, central and southern zones of genetic differentiation within the vicinity of Terra Nova Bay that were separated by the Aviator and Campbell Glaciers (Figure 5A; Fanciulli et al., 2001). It was also suggested that the population at Apostrophe Island (API; northern zone) was comprised of individuals that had more recently migrated from the central zone. The first COI sequences (n = 4) for this species were collected from near the Mario Zuchelli station (C. Beard, pers. comm.) and were reported in Stevens et al. (2006b). Hawes et al. (2010) provided 54 sequences and 25 new haplotypes from fine-scale (~15 km2) sampling in the southern zone (Terra Nova Bay). Broader-scale sampling by Carapelli et al. (2017) provided a further 114 sequences from 11 sites, including 38 new haplotypes. Each of these three genetic studies provided additional evidence to support the division of C. terranovus into the three zones as first suggested by Fanciulli et al. (2001) and our AMOVA analyses found that 59.8% of the sequence variation occurred between the three zones (Table 5; p < 0.01).
We provide a further five sequences (no new haplotypes) from near Jang Bogo Station in Terra Nova Bay, and O'Kane Glacier (Table 2; Tables S2, S6). We analyzed a total of 173 COI sequences for C. terranovus and identified 68 unique haplotypes from the 577 bp alignment. Sequences were grouped into the four distinct clades that have been previously identified for this species (Hawes et al., 2010; Carapelli et al., 2017). Overall, each clade contained between 20 and 78 sequences, mean sequence divergences within each clade ranged from 0.48 to 2.09%, and mean sequence divergences between clades ranged from 6.9 to 14.7% (Figure 5; Table 4; Table S6).
Southern Victoria Land Taxa
This region extends from the Drygalski Ice Tongue to the north and the Koettlitz Glacier to the south, and includes the Mackay Glacier region as well as the Victoria Land Dry Valleys including Victoria, Wright, Taylor, Garwood, Marshall, and Miers Valleys (Figures 1, 2). We analyzed a total of 1,023 COI sequences from the three springtail species that are currently known from this region; Antarcticinella monoculata (AMON; n = 33), Cryptopygus nivicolus (CNIV; n = 40), and Gomphiocephalus hodgsoni (GHOD; n = 950). All three species are range-restricted, with genetic differentiation found among different geographic locations. However, patterns of population structure vary among the three species. For example, individuals of G. hodgsoni at Mount Gran (n = 3) showed sequence divergences of 7.6% (Bennett et al., 2016), while individuals of C. nivicolus from Mount Gran (n = 7) were similar to those from other nearby sites such as Mount Seuss and Mount England (Beet et al., 2016). No sites were found with all three species present. Both C. nivicolus and A. monoculata were found at Springtail Point, while C. nivicolus and G. hodgsoni were both found at 5 sites (Towle Glacier, Tiger Island, and Mounts Seuss, Gran and England).
Antarcticinella monoculata (Salmon, 1965)
In total, 33 COI sequences were available for A. monoculata (Beet et al., 2016; Bennett et al., 2016) and all specimens were collected from within a 250 km2 range of the Mackay Glacier (Figure 6A; Table 3; Table S7). Sequences from the two northern-most sites, Mount Murray and Cliff Nunatak, were all the same haplotype (AMON-005) and were divergent from the other four haplotypes that were found at the more southerly sites (10.1% mean p-distance). An AMOVA revealed that 98.7% of the genetic variation occurred between these two geographically isolated populations (Table 5; p < 0.01).
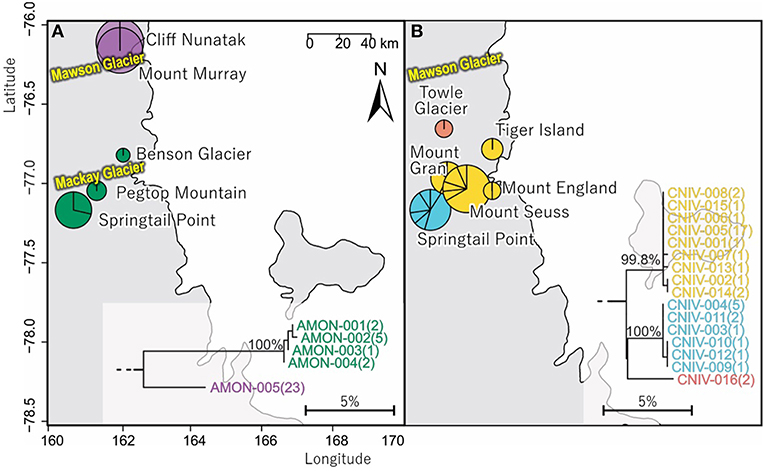
Figure 6. Geographical distribution within the vicinity and to the north of Mackay Glacier in southern Victoria Land of (A) the two distinct (10.1% mean divergence) lineages of Antarcticinella monoculata (AMON; n = 33; 530 bp), and (B) the three distinct (3.3–4.3% mean divergence) lineages of Cryptopygus nivicolus (CNIV; n = 40; 586 bp), highlighting differing population genetic structure for the two springtail species. Pie charts are proportionate to the number of sequences, centered at collection site co-ordinates.
Cryptopygus nivicolus (Salmon, 1965)
The first COI sequences (n = 2) for this species were obtained from Mount England (Stevens et al., 2006b). More recently, Bennett et al. (2016) and Beet et al. (2016) contributed another 32 sequences from an additional 5 sites, showing genetic divergence among habitats.
A further 6 sequences were included as part of our study (Tables 3, 4; Tables S2, S7). In total, we identified 16 haplotypes from the 40 COI sequences for this species, clustering into three distinct groups (3.3–4.3% mean divergence) corresponding to different geographic locations (Figure 6B). Springtail Point and the Towle Glacier site each had unique C. nivicolus sequences that were not found at any other site. The remaining nine haplotypes (n = 27 sequences) occurred across four sites in the vicinity of the Mackay Glacier (Mounts Gran, Seuss, England and Tiger Island; Figure 6B), suggesting individuals may have recently dispersed among hese sites (within 40 km of each other). Possible explanations for connectivity among these sites include dispersal via meltwater streams or in marine, nearshore environments when sea ice is absent along coastal boundaries. An AMOVA found that when sites at Springtail Point and Towle Glacier were designated as separate regions, only 35% of the genetic variation occurred between each of the three regions (Table 5; p < 0.01). However, genetically divergent populations of C. nivicolus were found at Springtail Point and the Towle Glacier site (Figure 6B), highlighting the importance of conservation strategies to prevent human-mediated transport of genetic variants between these currently isolated populations.
Gomphiocephalus hodgsoni (Carpenter, 1908)
Previous Studies
The first COI sequences for G. hodgsoni were contributed by a study that also examined allozymes and was focused on phylogeography (Stevens and Hogg, 2003). COI sequences were obtained for 45 individuals from 21 localities, and 14 unique haplotypes were identified (A-N). Among their findings it was suggested that two sympatric populations were present in Taylor Valley (1.5% divergence), and that recent transport (possibly by birds or humans) had occurred between Ross Island and Granite Harbor as similar haplotypes were shared between these locations. Sequences from Stevens and Hogg (2003) were included in their subsequent study (Stevens and Hogg, 2006), where genetically divergent populations were identified at Beaufort Island and in Taylor Valley, when compared to the remaining sites on Ross Island (n = 5 sites) and the continental mainland (n = 11 sites, including Taylor Valley). However, the Beaufort Island haplotype (our haplotype GHOD-010) has since been found at additional sites on the continent (see Table S9), while the unique Taylor Valley haplotype (our haplotype GHOD-011) remains restricted to Taylor Valley and belongs to Group Y in Collins and Hogg (2015) and Nolan et al. (2006).
A following study of G. hodgsoni (Nolan et al., 2006) targeted the two sympatric groups in Taylor Valley that had previously been identified by Stevens and Hogg (2003). The eight Taylor Valley sequences from Stevens and Hogg (2003) were also incorporated into their dataset, and 10 unique haplotypes (A–J) were identified from the combined dataset (n = 48). The study concluded that the two sympatric phylogroups (haplotypes A–F = group X; G–J = group Y) probably diverged less than 1 million years ago, around the time when ancient Lake Washburn flooded the Valley (40 kya). Ancestors of the two populations probably survived in isolated refugia at either end of the Valley (group Y inland and group X in higher elevation coastal areas), recolonizing the Valley and reconnecting following the retreat of Lake Washburn.
An additional five COI sequences and two new haplotypes were recovered from GenBank (McGaughran et al., 2008). Unfortunately, we were unable to determine which sequences corresponded to haplotype codes referred to in the manuscript. As these sequences were included in the subsequent study of McGaughran et al. (2010), they have also been included in our study. The McGaughran et al. (2008) study included sequences from Stevens and Hogg (2003) and identified 20 haplotypes (G1-G20) from a total dataset of 96 sequences from the wider McMurdo Dry Valleys and Ross Island area. Divergences of G. hodgsoni COI sequences were up to 2.1%, suggesting the populations had diverged within the last million years (based on molecular clock calibration of 1.5–2.3%/my e.g., Brower, 1994; Juan et al., 1996; Quek et al., 2004). Further, haplotype diversity tended to be higher at more inland sites and particularly at higher altitudes, suggesting this was a result of persistence in refugial habitats. McGaughran et al. (2008) also suggested that Ross Island individuals were derived from a founder population that originated from the Dry Valleys.
McGaughran et al. (2010) compared patterns of sequence divergences of G. hodgsoni to those of Cryptopygus antarcticus from the Antarctic Peninsula and provided COI and COII sequences for both taxa. Existing G. hodgsoni COI sequences from Stevens and Hogg (2003) and McGaughran et al. (2008) were also included, and 45 haplotypes (H1–H45) were identified from their final alignment of 289 individuals (471 bp). In reviewing these sequences, we were able to identify exact site information for all but 31 specimens (see Table S9). Key findings from McGaughran et al. (2010) included limited haplotype sharing between local sites within both the Peninsula and continental areas, and that survival in refugial habitats was likely to have occurred on a Pleistocene timescale. However, the Peninsula contained more rare haplotypes and it was suggested that differences in population structure were a result of landscape differences and colonization events between the two localities. Greenslade et al. (2011) provided three additional COI sequences (658 bp) for the existing haplotype H43 from McGaughran et al. (2010) (our haplotype GHOD-046) in a study which confirmed Gomphiocephalus as a distinct genus.
A further 90 G. hodgsoni COI sequences were contributed by Demetras (2010) from the southern Dry Valleys area (Marshall, Garwood and Miers Valleys) with limited diversity found (10 haplotypes TABS Gh1—Gh10; <0.8% divergence). Based on a haplotype network analysis, it was suggested that all 10 haplotypes were derived from a single lineage (300–500 kya). Possible reasons for the low diversity in G. hodgsoni from the southern Dry Valleys include more recent recolonization of this area or bottleneck events and differences in landscapes as compared to the larger Dry Valleys (e.g., Taylor) where there is much higher COI diversity. As the data from Demetras (2010) were previously unpublished, we have now uploaded them to BOLD and added them to our analyses.
Collins and Hogg (2015) provided a further 151 G. hodgsoni COI sequences as part of a two-hourly time-series of pitfall trap collections from Spaulding Pond in Taylor Valley. This study targeted the two main “X” and “Y” haplotype groupings previously reported by Nolan et al. (2006), known to occur in sympatry at Spaulding Pond (Gh1-Gh12 = Y; Gh13-Gh19 = X). More individuals were found from group Y (n = 120) relative to group X (n = 31). As group Y is thought to have recolonized from inland “colder” refugial sites, this study inferred that the site was dominated by intrinsically cold-adapted individuals, and that the relative proportions of X and Y individuals could change with a warming climate. Furthermore, activity of individuals from the X and Y haplotype lineages during each two-hourly pitfall trap collection was more closely linked to air temperature than any of the other measured environmental variables. This highlights the potential for temporal shifts in the genetic structure of populations as a consequence of environmental changes.
An additional 67 sequences (658 bp; 16 haplotypes, 8 new) were contributed by Bennett et al. (2016), along with sequences from A. monoculata and C. nivicolus. This study was focused on past isolation events using a molecular clock analysis. They concluded that isolation was likely to have occurred during the last 4 million years and that glaciation events since that time have further contributed to the high COI diversity within both G. hodgsoni and C. nivicolus. In particular, specimens of G. hodgsoni from Mount Gran (our haplotype GHOD-071) were found to be highly divergent (>7%) from the remaining sampled dataset. In a study focused on genetic diversity of Collembola from sites in the vicinity of Mackay Glacier and further north, Beet et al. (2016) contributed 66 new sequences (527 bp; 5 new haplotypes). Sequences from Bennett et al. (2016) were also included in Beet et al. (2016) analyses and, based on the combined dataset, it was hypothesized that populations may have been isolated for 3–5 million years, consistent with a collapse and reformation of the West Antarctic Ice Sheet.
Our Findings
As the most widespread and well-studied springtail in the Ross Sea region, the dataset of COI sequences for G. hodgsoni (n = 950) was larger than that of any other species and represented specimens from 67 sites (Tables 3, 4; Tables S8, S9). Overall, 620 sequences were available from nine published studies as outlined above, and a further 330 previously unpublished sequences have now been added here (Table S2). The majority of sequences were <2.6% divergent, while distinct populations were present at Towle Glacier (n = 6; 5.9% divergence) and Mount Gran (n = 3; 7.3% divergence) which are located further inland (Figure 7). Similarly, genetically divergent populations of C. nivicolus were present at the inland sites Towle Glacier and Springtail Point, suggesting that greater connectivity occurs among coastal sites. Overall, an AMOVA revealed that the majority (>60%) of haplotype diversity occurred within regions (Table 5; p < 0.01), particularly as the highly diverged individuals from sites Towle Glacier and Mount Gran were sequenced at low numbers.
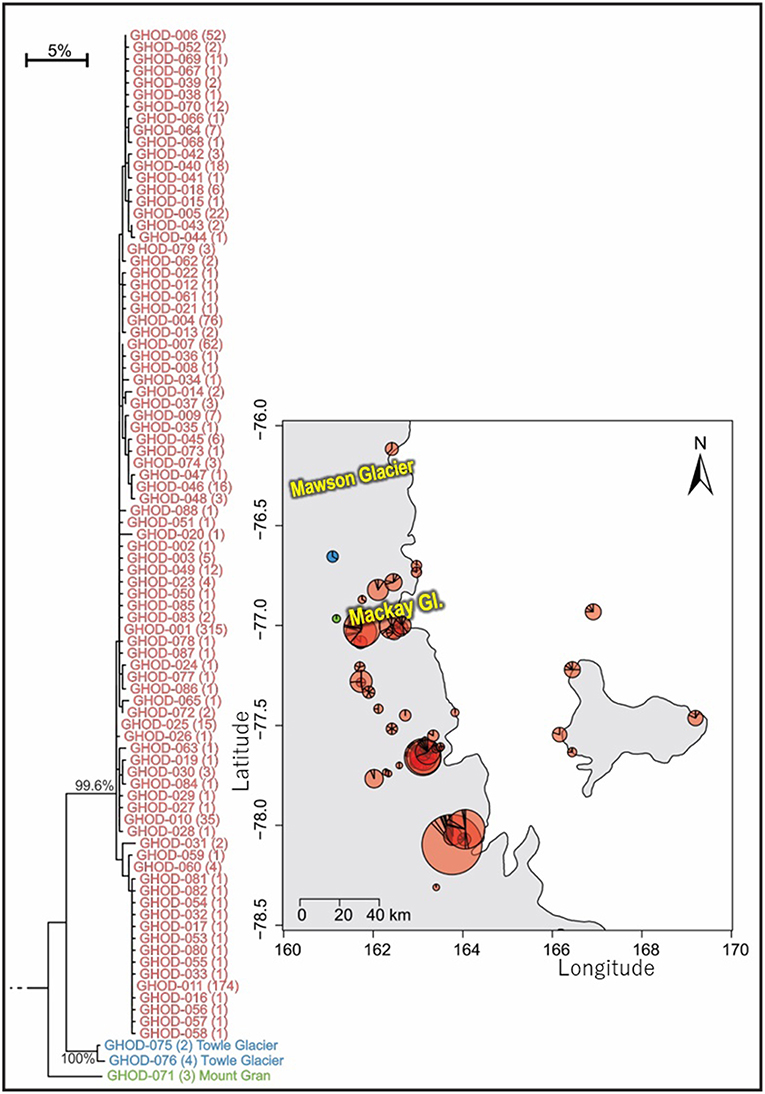
Figure 7. Geographical distribution of the three distinct (5.9–7.3% mean divergence) lineages of Gomphiocephalus hodgsoni within southern Victoria Land for specimens where collection data were available (GHOD; n = 904; 422 bp), showing widespread distribution (500 km reach) of the main group of haplotypes (n = 895; 2.6% mean divergence) and distinct populations at both Mount Gran and Towle Glacier sites, further inland. Pie charts are proportionate to the number of sequences, centered at collection site co-ordinates. See Tables S8, S9 for further haplotype and collection details for each specimen.
Tripp Island is approximately 70 km from other sites and located to the north of Mawson glacier which appeared to be a dispersal barrier for A. monoculata (>10% divergence). However, individuals of G. hodgsoni from Tripp Island were not genetically divergent (<0.5%) relative to other locations. In contrast, individuals of G. hodgsoni at Mount Gran <10 km from other sites (Figure 7) were highly genetically divergent (>7%). These findings highlight that geographical barriers to dispersal such as glaciers, and lack of water transport between locations, which is unlikely for sites further inland, have a stronger influence on the population genetic structures of Antarctic springtails than distance alone.
Concluding Discussion
In this study we examined the available COI sequence data for all seven of the Collembola species that occur within Victoria Land (71° to 78.5°S). All species harbored distinct lineages (1.4–14.7% mean sequence divergences) that were isolated by geological or glaciological features, rather than by distance alone. Such levels of COI diversity are suggestive of long-term isolation and lack of dispersal among locations.
In several cases, the distribution of genetic variants among sites differed for the different species. Indeed, differing distributional patterns were previously reported in a large-scale study of the three species C. terranovus, F. grisea, and G. hodgsoni (Caruso et al., 2009). Here, we showed that sequence divergence for F. grisea (0.2%) and C. terranovus (10.8%) differed greatly among the same sites in northern Victoria Land. As a possible explanation, F. grisea may have more recently dispersed throughout this area whereas C. terranovus may have remained isolated by the Aviator and Campbell Glaciers (e.g., Fanciulli et al., 2001; Carapelli et al., 2017). In southern Victoria Land (SVL), Springtail Point harbored a distinct lineage (>3.3% divergence) of C. nivicolus whereas A. monoculata from Springtail Point was highly similar (<0.9% divergence) to individuals from nearby sites. Similarly, the Mount Gran population of G. hodgsoni was highly distinct (7.3% divergence) whereas sequences of C. nivicolus from Mount Gran were identical to those from other sites. Populations to the north of Mawson Glacier were genetically distinct for A. monoculata (>10% sequence divergence; Cliff Nunatak and Mount Murray) while the G. hodgsoni population (Tripp Island) was genetically similar (0.5%), even though these sites are geographically isolated from other sites by >70 km (Figures 2, 6, 7).
Given the high levels of genetic divergence among populations for all seven springtail species (1.7–14.7%), we highlight the potential that these distinct lineages could be cryptic species. Cryptic diversity has been suggested for C. terranovus in NVL (Hawes et al., 2010; Carapelli et al., 2017) and G. hodgsoni, C. nivicolus, and A. monoculata in SVL (Beet et al., 2016). Suggestions of cryptic diversity for the springtail F. grisea between NVL and the Antarctic Peninsula (Torricelli et al., 2010a,b) have now been validated through morphological differences and redescription of the species (Greenslade, 2018). However, the potential for cryptic variability among Victoria Land specimens of F. grisea has not previously been suggested, although the Cape Hallett population has been identified as genetically distinct, possibly resulting from pre-Pleistocene isolation (Torricelli et al., 2010b).
The Tucker Glacier in NVL provides an example of a small-scale (< 16 km) barrier to springtail dispersal as genetically distinct (>5% sequence divergence) populations of both C. cisantarcticus and F. grisea have been found either side of the glacier. Previous studies using the COII gene have also demonstrated this for K. klovstadi (Frati et al., 2001; Stevens et al., 2007), suggesting the potential for microspeciation processes occurring at these sites. In SVL, we show that the phylogeographic patterns differed for each of the three species, and we highlight Springtail Point, Towle Glacier, Mount Gran and sites to the north of Mawson Glacier as possible sites of conservation focus, as these locations harbored genetically divergent (3.3–10.1%) populations. The vicinity of the Drygalski Ice Tongue continues to provide a current barrier to dispersal for springtail taxa between NVL and SVL and we found no evidence of gene flow or species sharing between these regions.
Future studies focused on longer COI sequences (or indeed, additional genetic markers), are likely to reveal further genetic diversity for springtails in Victoria Land. The potential for temporal changes in genetic diversity as a consequence of environmental changes such as climate warming has also been highlighted. Next generation sequencing and metabarcoding approaches for genomic monitoring will be important for future studies, particularly to detect spatial and temporal shifts in genetic diversity. With a warming climate, future glacier melt will provide additional opportunities for the dispersal of springtails (via flotation) while also exposing new habitats for colonization.
The Antarctic Conservation Biogeographic Regions (Terauds et al., 2012; Terauds and Lee, 2016) provide a geographical framework for assessing broad-scale biological diversity. However, we suggest that knowledge of local-scale patterns of genetic diversity will be critical for addressing ecological and physiological questions, such as those pertaining to dispersal, and responses to climate changes. The understanding of relevant temporal and spatial scales as well as the current distribution of genetic diversity is essential to identify current barriers to dispersal as well as sites that harbor unique genetic resources and, therefore, areas of conservation value.
Data Availability
All previously unpublished sequences and associated data are available in the dataset “DSANTSP18” on BOLD. GenBank accession numbers and BOLD IDs are provided in supplementary data sheets for all sequences included in this manuscript. Original trace files can be found associated with specimens in the BOLD dataset, where available. For any additional information, contact GC: gec9@students.waikato.ac.nz/.
Author Contributions
GC and IH conceived the study. IH collected new specimens from NVL, and processed these with GC. GC and IH collected and processed additional specimens from SVL. All alignments and analyses were performed by GC, with assistance from AB using R. The manuscript was prepared by GC, with contributions from all co-authors.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer, BV, declared past collaborations with one of the authors, PC, to the handling Editor.
Acknowledgments
We acknowledge support from a Waikato University Doctoral Scholarship, a Waikato Graduate Women Merit Award for Doctoral Study and an Antarctica New Zealand Postgraduate Scholarship (Sir Robin Irvine) to GC. IH is grateful to Antarctica New Zealand, Craig Cary and the Korean Polar Research Institute (KOPRI) for arranging/providing logistic support for field work in NVL, and to Polar Knowledge Canada for financial support. PC was supported by NERC core funding to the British Antarctic Survey's ‘Biodiversity, Evolution and Adaptation’ Team. We are also grateful for support provided to the Canadian Centre for DNA Barcoding from Genome Canada and the Ontario Genomics Institute as part of the International Barcode of Life Project and thank the Ontario Ministry of Economic Development and Innovation for support of the BOLD database. This paper contributes to the ‘State of the Antarctic Ecosystem’ (AntEco) and the Antarctic Thresholds, Ecosystems Resilience and Adaptation (AnT-ERA) programmes of SCAR. The satellite imagery used to generate Figures 1, 2 was retrieved from GloVis, courtesy of the NASA EOSDIS Land Processes Distributed Active Archive Center (LP DAAC), USGS/Earth Resources Observation and Science (EROS) Center, Sioux Falls, South Dakota, (https://lpdaac.usgs.gov/data_access/glovis).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2019.00076/full#supplementary-material
References
Adams, B. J., Bardgett, R. D., Ayres, E., Wall, D. H., Aislabie, J., Bamforth, S., et al. (2006). Diversity and distribution of Victoria Land biota. Soil Biol. Biochem. 38, 3003–3018. doi: 10.1016/j.soilbio.2006.04.030
Becker, R., Wilks, A., and Brownrigg, R. (2016). Mapdata: Extra Map Databases. R package version 2.2–6.
Beet, C. R., Hogg, I. D., Collins, G. E., Cowan, D. A., Wall, D. H., and Adams, B. J. (2016). Genetic diversity among populations of Antarctic springtails (Collembola) within the mackay glacier ecotone. Genome 59, 762–770. doi: 10.1139/gen-2015-0194
Bennett, K. R., Hogg, I. D., Adams, B. J., and Hebert, P. D. N. (2016). High levels of intraspecific genetic divergences revealed for Antarctic springtails: evidence for small-scale isolation during pleistocene glaciation. Biol. J. Linn. Soc. 119, 166–178. doi: 10.1111/bij.12796
Bergstrom, D. M., and Chown, S. L. (1999). Life at the front: history, ecology and change on southern ocean islands. Trends Ecol. Evol. 14, 472–477. doi: 10.1016/S0169-5347(99)01688-2
Brower, A. V. (1994). Rapid morphological radiation and convergence among races of the butterfly Heliconius erato inferred from patterns of mitochondrial DNA evolution. Proc. Natl. Acad. Sci. U.S.A. 91, 6491–6495. doi: 10.1073/pnas.91.14.6491
Carapelli, A., Leo, C., and Frati, F. (2017). High levels of genetic structuring in the antarctic springtail cryptopygus terranovus. Antarct. Sci. 29, 311–323. doi: 10.1017/S0954102016000730
Carpenter, G. H. (1902). “Insecta: Aptera - Collembola,” in Report on the Collections of Natural History Made in the Antarctic Regions During the Voyage of the Southern Cross, ed. E. R. Lankester (London: British Museum), 221–223.
Carpenter, G. H. (1908). “Arthropoda: Insecta - Aptera,” in National Antarctic Expedition 1901-1904 Natural History, ed. E. R. Lankester (London, UK: British Museum), 1–6.
Caruso, T., Hogg, I. D., Carapelli, A., Frati, F., and Bargagli, R. (2009). Large-scale spatial patterns in the distribution of collembola (Hexapoda) species in Antarctic terrestrial ecosystems. J. Biogeogr. 36, 879–886. doi: 10.1111/j.1365-2699.2008.02058.x
Chown, S. L., and Convey, P. (2007). Spatial and temporal variability across life's hierarchies in the terrestrial Antarctic. Philos. Trans. R. Soc. B-Biol. Sci. 362, 2307–2331. doi: 10.1098/rstb.2006.1949
Colesie, C., Green, T. G. A., Türk, R., Hogg, I. D., Sancho, L. G., and Büdel, B. (2014). Terrestrial biodiversity along the ross sea coastline, Antarctica: lack of a latitudinal gradient and potential limits of bioclimatic modeling. Polar Biol. 37, 1197–1208. doi: 10.1007/s00300-014-1513-y
Collins, G. E., and Hogg, I. D. (2015). Temperature-related activity of Gomphiocephalus hodgsoni (Collembola) mitochondrial DNA (COI) haplotypes in Taylor Valley, Antarctica. Polar Biol. 39, 1–11. doi: 10.1007/s00300-015-1788-7
Convey, P. (2013). “Antarctic ecosystems,” in Encyclopedia of Biodiversity, 2nd Edn (Elsevier Inc.), 179–188. doi: 10.1016/B978-0-12-384719-5.00264-1
Convey, P., Gibson, J. A., Hillenbrand, C.-D., Hodgson, D. A., Pugh, P. J. A., Smellie, J. L., et al. (2008). Antarctic terrestrial life – challenging the history of the frozen continent? Biol. Rev. 83, 103–117. doi: 10.1111/j.1469-185X.2008.00034.x
Darriba, D., Taboada, G. L., Doallo, R., and Posada, D. (2012). jModelTest 2: more models, new heuristics and parallel computing. Nat. Methods 9:772. doi: 10.1038/nmeth.2109
Demetras, N. J. (2010). Phylogeography and Genetic Diversity of Terrestrial Arthropods From the Ross Dependency, Antarctica. MSc Thesis, University of Waikato, New Zealand.
Excoffier, L., Smouse, P. E., and Quattro, J. M. (1992). Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131, 479–491.
Fanciulli, P. P., Summa, D., Dallai, R., and Frati, F. (2001). High levels of genetic variability and population differentiation in Gressittacantha terranova (Collembola, Hexapoda) from Victoria Land, Antarctica. Antarct. Sci. 13, 246–254. doi: 10.1017/S0954102001000360
Frati, F., and Dell'Ampio, E. (2000). Molecular phylogeny of three subfamilies of the neanuridae (Insecta, Collembola) and the position of the antarctic species Friesea grisea Schäffer. Pedobiologia 44, 342–360. doi: 10.1078/S0031-4056(04)70054-2
Frati, F., Fanciulli, P. P., Carapelli, A., De Carlo, L., and Dallai, R. (1996). Collembola of northern Victoria Land: distribution, population structure and preliminary molecular data to study origin and evolution of Antarctic Collembola. in Proceedings of the 3rd Meeting on Antarctic Biology, eds G. di Prisco, S. Focardi and P. Luporini (Santa Margherita Ligure: Camerino Univ. Press), 321–330.
Frati, F., Spinsanti, G., and Dallai, R. (2001). Genetic variation of mtCOII gene sequences in the collembolan Isotoma klovstadi from Victoria Land, Antarctica: evidence for population differentiation. Polar Biol. 24, 934–940. doi: 10.1007/s003000100302
Freckman, D. W., and Virginia, R. A. (1993). Extraction of nematodes from dry valley Antarctic soils. Polar Biol. 13, 483–487. doi: 10.1007/BF00233139
Green, T. G. A., Sancho, L. G., Türk, R., Seppelt, R. D., and Hogg, I. D. (2011). High diversity of lichens at 84°S, queen maud mountains, suggests preglacial survival of species in the ross sea region, Antarctica. Polar Biol. 34, 1211–1220. doi: 10.1007/s00300-011-0982-5
Greenslade, P. (2015). Synonymy of two monobasic anurophorinae genera (Collembola: Isotomidae) from the Antarctic continent. N. Zeal. Entomol. 38, 134–141. doi: 10.1080/00779962.2015.1033810
Greenslade, P. (2018). An Antarctic biogeographical anomaly resolved: the true identity of a widespread species of Collembola. Polar Biol. 41, 969–981. doi: 10.1007/s00300-018-2261-1
Greenslade, P., Stevens, M. I., Torricelli, G., and D'Haese, C. A. (2011). An ancient Antarctic endemic genus restored: morphological and molecular support for Gomphiocephalus hodgsoni (Collembola: Hypogastruridae). Syst. Entomol. 36, 223–240. doi: 10.1111/j.1365-3113.2010.00553.x
Gregory, J. W. (1909). Further antarctic reports of the expedition of 1901-1904. Geogr. J. 34, 290–297. doi: 10.2307/1777145
Gressitt, J. L., and Leech, R. E. (1961). Insect habitats in Antarctica. Polar Rec. 10, 501–504. doi: 10.1017/S0032247400051871
Gressitt, J. L., Leech, T. S., and Wise, K. A. J. (1963). Entomological investigations in Antarctica. Pac. Insects 5, 287–304.
Guindon, S., and Gascuel, O. (2003). A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52, 696–704. doi: 10.1080/10635150390235520
Hawes, T. C., Torricelli, G., and Stevens, M. I. (2010). Haplotype diversity in the Antarctic springtail Gressittacantha terranova at fine spatial scales-a holocene twist to a pliocene tale. Antarct. Sci. 22, 766–773. doi: 10.1017/S0954102010000490
Hawes, T. C., Worland, M. R., Bale, J. S., and Convey, P. (2008). Rafting in Antarctic Collembola. J. Zool. 274, 44–50. doi: 10.1111/j.1469-7998.2007.00355.x
Janetschek, H. (1963). On the terrestrial fauna of the ross-sea area, Antarctica (Preliminary report). Pacific Insects 5, 305–311.
Janetschek, H. (1967). Growth and maturity of the springtail, Gomphiocephalus hodgsoni Carpenter, from South Victoria Land and Ross Island. Entomol. Antarct. 10, 295–305. doi: 10.1029/AR010p0295
Janetschek, H. (1970). “Environments and ecology of terrestrial arthropods in the high Antarctic,” in Antarctic Ecology, ed. M. Holdgate (London: Academic Press), 871–885.
Juan, C., Oromi, P., and Hewitt, G. M. (1996). Phylogeny of the genus hegeter (Tenebrionidae, Coleoptera) and its colonization of the canary islands deduced from cytochrome oxidase i mitochondrial DNA sequences. Heredity 76, 392–403. doi: 10.1038/hdy.1996.57
Kamvar, Z. N., Brooks, J. C., and Grünwald, N. J. (2015). Novel R tools for analysis of genome-wide population genetic data with emphasis on clonality. Front. Genet. 6:208. doi: 10.3389/fgene.2015.00208
Kamvar, Z. N., Tabima, J. F., and Grünwald, N. J. (2014). Poppr: an R package for genetic analysis of populations with clonal, partially clonal, and/or sexual reproduction. PeerJ 2:e281. doi: 10.7717/peerj.281
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Lee, J. R., Raymond, B., Bracegirdle, T. J., Chadès, I., Fuller, R. A., Shaw, J. D., et al. (2017). Climate change drives expansion of Antarctic ice-free habitat. Nature 547, 49–54. doi: 10.1038/nature22996
Macnamara, C. (1919). Popular and practical entomology. further remarks on collembola. Can. Entomol. 51, 241–246. doi: 10.4039/Ent51241-11
McGaughran, A., Hogg, I. D., and Convey, P. (2011a). Extended ecophysiological analysis of Gomphiocephalus hodgsoni (Collembola): flexibility in life history strategy and population response. Polar Biol. 34, 1713–1725. doi: 10.1007/s00300-011-1001-6
McGaughran, A., Hogg, I. D., and Stevens, M. I. (2008). Patterns of population genetic structure for springtails and mites in southern Victoria Land, Antarctica. Mol. Phylogenet. Evol. 46, 606–618. doi: 10.1016/j.ympev.2007.10.003
McGaughran, A., Stevens, M. I., Hogg, I. D., and Carapelli, A. (2011b). Extreme glacial legacies: a synthesis of the Antarctic springtail phylogeographic record. Insects 2, 62–82. doi: 10.3390/insects2020062
McGaughran, A., Torricelli, G., Carapelli, A., Frati, F., Stevens, M. I., Convey, P., et al. (2010). Contrasting phylogeographical patterns for springtails reflect different evolutionary histories between the Antarctic Peninsula and continental Antarctica. J. Biogeogr. 37, 103–119. doi: 10.1111/j.1365-2699.2009.02178.x
Nardi, F., Spinsanti, G., Boore, J. L., Carapelli, A., Dallai, R., and Frati, F. (2003). Hexapod origins: monophyletic or paraphyletic? Science 299, 1887–1889. doi: 10.1126/science.1078607
Nolan, L., Hogg, I. D., Stevens, M. I., and Haase, M. (2006). Fine scale distribution of mtDNA haplotypes for the springtail Gomphiocephalus hodgsoni (Collembola) corresponds to an ancient shoreline in Taylor Valley, continental Antarctica. Polar Biol. 29, 813–819. doi: 10.1007/s00300-006-0119-4
Peat, H. J., Clarke, A., and Convey, P. (2007). Diversity and biogeography of the Antarctic flora. J. Biogeogr. 34, 132–146. doi: 10.1111/j.1365-2699.2006.01565.x
Quek, S. P., Davies, S. J., Itino, T., and Pierce, N. E. (2004). Codiversification in an ant-plant mutualism: stem texture and the evolution of host use in Crematogaster (Formicidae: Myrmicinae) inhabitants of Macaranga (Euphorbiaceae). Evolution (N. Y). 58, 554–570. doi: 10.1111/j.0014-3820.2004.tb01678.x
Salmon, J. T. (1962). A new species and redescriptions of Collembola from Antarctica. Pacific Insects 4, 887–894.
Schaeffer, C. (1891). Die Collembolen von Sud-Georgien Nach der Ausbeute der Deutschen Station von 1882/83. Jahrbuch der Hamburgischen Wissenschaftlichen Anstalten 9, 193–201.
Sinclair, B. J., and Sjursen, H. (2001). Cold tolerance of the Antarctic springtail Gomphiocephalus hodgsoni (Collembola, Hypogastruridae). Antarct. Sci. 13, 271–279. doi: 10.1017/S0954102001000384
Sinclair, B. J., and Stevens, M. I. (2006). Terrestrial microarthropods of Victoria Land and Queen Maud Mountains, Antarctica: implications of climate change. Soil Biol. Biochem. 38, 3158–3170. doi: 10.1016/j.soilbio.2005.11.035
Stevens, M. I., and D'Haese, C. A. (2016). Morphologically tortured: taxonomic placement of an Antarctic springtail (Collembola: Isotomidae) misguided by morphology and ecology. Zool. Scr. 46, 180–187. doi: 10.1111/zsc.12204
Stevens, M. I., Fjellberg, A., Greenslade, P., Hogg, I. D., and Sunnucks, P. (2006a). Redescription of the Antarctic springtail Desoria klovstadi using morphological and molecular evidence. Polar Biol. 29, 820–830. doi: 10.1007/s00300-006-0120-y
Stevens, M. I., Frati, F., McGaughran, A., Spinsanti, G., and Hogg, I. D. (2007). Phylogeographic structure suggests multiple glacial refugia in northern Victoria Land for the endemic Antarctic springtail Desoria klovstadi (Collembola, Isotomidae). Zool. Scr. 36, 201–212. doi: 10.1111/j.1463-6409.2006.00271.x
Stevens, M. I., Greenslade, P., Hogg, I. D., and Sunnucks, P. (2006b). Southern hemisphere springtails: could any have survived glaciation of Antarctica? Mol. Biol. Evol. 23, 874–882. doi: 10.1093/molbev/msj073
Stevens, M. I., and Hogg, I. D. (2002). Expanded distributional records of Collembola and Acari in southern Victoria land, Antarctica. Pedobiologia (Jena). 46, 485–495. doi: 10.1078/0031-4056-00154
Stevens, M. I., and Hogg, I. D. (2003). Long-term isolation and recent range expansion from glacial refugia revealed for the endemic springtail Gomphiocephalus hodgsoni from Victoria Land, Antarctica. Mol. Ecol. 12, 2357–2369. doi: 10.1046/j.1365-294X.2003.01907.x
Stevens, M. I., and Hogg, I. D. (2006). Contrasting levels of mitochondrial DNA variability between mites (Penthalodidae) and springtails (Hypogastruridae) from the trans-antarctic mountains suggest long-term effects of glaciation and life history on substitution rates, and speciation processes. Soil Biol. Biochem. 38, 3171–3180. doi: 10.1016/j.soilbio.2006.01.009
Terauds, A., Chown, S. L., Morgan, F., Peat, H. J., Watts, D. J., Keys, H., et al. (2012). Conservation biogeography of the Antarctic. Divers. Distrib. 18, 726–741. doi: 10.1111/j.1472-4642.2012.00925.x
Terauds, A., and Lee, J. R. (2016). Antarctic biogeography revisited: updating the antarctic conservation biogeographic regions. Divers. Distrib. 22, 836–840. doi: 10.1111/ddi.12453
Torricelli, G., Carapelli, A., Convey, P., Nardi, F., Boore, J. L., and Frati, F. (2010a). High divergence across the whole mitochondrial genome in the “pan-Antarctic” springtail Friesea grisea: evidence for cryptic species? Gene 449, 30–40. doi: 10.1016/j.gene.2009.09.006
Torricelli, G., Frati, F., Convey, P., Telford, M., and Carapelli, A. (2010b). Population structure of Friesea grisea (Collembola, Neanuridae) in the Antarctic Peninsula and Victoria Land: evidence for local genetic differentiation of pre-Pleistocene origin. Antarct. Sci. 22, 757–765. doi: 10.1017/S0954102010000775
Willem, V. (1902). “Collemboles,” in Résultats du voyage du S.Y. Belgica en 1897-1898-1899: sous le commandement de A. de Gerlache de Gomery. Rapports scientifiques publiés aux frais du gouvernement belge, sous la direction de la Commission de la Belgica. Anvers: Impr. J.E. Buschmann, 1901–1910. doi: 10.5962/bhl.title.2170
Wise, K. A. J. (1967). Collembola (springtails). Antarct. Res. Ser. 10, 123–148. doi: 10.1029/AR010p0123
Keywords: Antarctica, biogeography, collembola, dispersal, mitochondrial DNA barcodes, population genetic structure, species diversity, springtails
Citation: Collins GE, Hogg ID, Convey P, Barnes AD and McDonald IR (2019) Spatial and Temporal Scales Matter When Assessing the Species and Genetic Diversity of Springtails (Collembola) in Antarctica. Front. Ecol. Evol. 7:76. doi: 10.3389/fevo.2019.00076
Received: 08 August 2018; Accepted: 28 February 2019;
Published: 22 March 2019.
Edited by:
Angela McGaughran, Australian National University, AustraliaReviewed by:
Bettine Van Vuuren, University of Johannesburg, South AfricaKaty Morgan, University of Bath, United Kingdom
Copyright © 2019 Collins, Hogg, Convey, Barnes and McDonald. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ian D. Hogg, ian.hogg@polar.gc.ca orcid.org/0000-0002-6685-0089
†Andrew Barnes orcid.org/0000-0002-6499-381X
 Gemma E. Collins
Gemma E. Collins Ian D. Hogg1,2*
Ian D. Hogg1,2*  Peter Convey
Peter Convey Ian R. McDonald
Ian R. McDonald