Abstract
RNA interference (RNAi) holds considerable promise as a therapeutic approach to silence disease-causing genes, particularly those that encode so-called ‘non-druggable’ targets that are not amenable to conventional therapeutics such as small molecules, proteins, or monoclonal antibodies. The main obstacle to achieving in vivo gene silencing by RNAi technologies is delivery. Here we show that chemically modified short interfering RNAs (siRNAs) can silence an endogenous gene encoding apolipoprotein B (apoB) after intravenous injection in mice. Administration of chemically modified siRNAs resulted in silencing of the apoB messenger RNA in liver and jejunum, decreased plasma levels of apoB protein, and reduced total cholesterol. We also show that these siRNAs can silence human apoB in a transgenic mouse model. In our in vivo study, the mechanism of action for the siRNAs was proven to occur through RNAi-mediated mRNA degradation, and we determined that cleavage of the apoB mRNA occurred specifically at the predicted site. These findings demonstrate the therapeutic potential of siRNAs for the treatment of disease.
Similar content being viewed by others
Main
RNAi has been applied widely as a target validation tool in post-genomic research, and it represents a potential strategy for in vivo target validation and therapeutic product development1. In vivo gene silencing with RNAi has been reported using both viral vector delivery2 and high-pressure, high-volume intravenous (i.v.) injection of synthetic siRNAs3, but these approaches have limited if any clinical use. In vivo gene silencing has also been reported after local, direct administration (intravitreal, intranasal and intrathecal) of siRNAs to sequestered anatomical sites in models of choroidal neovascularization4, lung ischaemia-reperfusion injury5 and neuropathic pain6, respectively. These reported approaches demonstrate the potential for delivery to organs such as the eye, lungs and central nervous system. However, there are no published reports of systemic activity for siRNAs towards endogenous targets after conventional and clinically acceptable routes of administration. A critical requirement for achieving systemic RNAi in vivo is the introduction of ‘drug-like’ properties, such as stability, cellular delivery and tissue bioavailability, into synthetic siRNAs.
Conferring drug-like properties on siRNAs
In exploring the potential of synthetic siRNAs to silence endogenous target genes, we found that chemically stabilized and cholesterol-conjugated siRNAs7 have markedly improved pharmacological properties in vitro and in vivo. Chemically stabilized siRNAs with partial phosphorothioate backbone and 2’-O-methyl sugar modifications on the sense and antisense strands showed significantly enhanced resistance towards degradation by exo- and endonucleases in serum and in tissue homogenates. The conjugation of cholesterol to the 3′ end of the sense strand of a siRNA molecule by means of a pyrrolidine linker (thereby generating chol-siRNA) did not result in a significant loss of gene-silencing activity in cell culture. Furthermore, unlike unconjugated siRNAs, a chol-siRNA directed to luciferase (chol-luc-siRNA) showed reduction in luciferase activity in HeLa cells transiently expressing luciferase, with a half-maximal inhibitory concentration (IC50) of about 200 nM in the absence of transfection reagents or electroporation.
Binding of chol-siRNAs to human serum albumin (HSA) was determined by surface plasmon resonance measurement (data not shown). Unconjugated siRNAs demonstrated no measurable binding to HSA, whereas chol-siRNAs bound to HSA with an estimated dissociation constant (Kd) of 1 µM. Presumably because of enhanced binding to serum proteins, chol-siRNAs administered to rats by i.v. injection showed improved in vivo pharmacokinetic properties as compared to unconjugated siRNAs. After i.v. injection in rats at 50 mg kg-1, radioactively labelled chol-siRNAs had an elimination half life (two compartments), t1/2 of 95 min and a corresponding plasma clearance (CL) of 0.5 ml min-1, whereas unconjugated siRNAs had a t1/2 of 6 min and CL of 17.6 ml min-1. As measured by an RNase protection assay (RPA), chol-siRNAs showed broad tissue biodistribution 24 h after injection in mice. Although no detectable amounts of unconjugated siRNAs were observed in tissue samples, significant levels of chol-siRNAs were detected in liver, heart, kidney, adipose, and lung tissue samples. Together, these studies demonstrate that cholesterol conjugation significantly improves in vivo pharmacological properties of siRNAs.
Selection of apoB as an endogenous gene target
Apolipoprotein B is the essential protein for formation of low-density lipoproteins (LDL) in metabolism of dietary and endogenous cholesterol, and is the ligand for the LDL receptor8. Mouse apoB is a large protein of 4,515 amino acids and is expressed predominantly in liver and jejunum. apoB mRNA is subject to post-transcriptional editing, and the unedited and edited transcripts encode the full-length protein apoB-100, and a carboxy-terminal truncated isoform, apoB-48, respectively. In mice, editing of apoB mRNA occurs in both the liver and jejunum: apoB-48 is the predominant protein form in the jejunum and both apoB-48 and apoB-100 are expressed in the liver. Heterozygous knockout mice for apoB show a 20% decrease in cholesterol levels and are resistant to diet-induced hypercholesterolaemia9.
Serum levels of apoB, LDL and cholesterol correlate significantly with increased risk of coronary artery disease (CAD). A diminished number of functional LDL receptors on the cell surface, disrupting receptor-mediated removal of apoB-containing LDL from circulation, has been identified as the basis for familial hypercholesterolaemia (FH)10. Patients with homozygous and heterozygous FH have accelerated CAD leading to premature atherosclerosis and cardiac mortality. Conversely, patients with hypobetalipoproteinaemia have reduced levels of LDL and cholesterol and are at reduced risk for CAD11. Accordingly, lowering of serum cholesterol and LDL levels is a predominant clinical strategy for management of CAD and is achieved by modification of dietary sources of cholesterol and/or inhibition of endogenous cholesterol synthesis with pharmacological therapies. Notwithstanding significant improvements in the management of CAD with these approaches, millions of patients remain at significant risk for CAD and its clinical sequelae—acute coronary syndromes such as myocardial infarction and cardiac mortality—due to advanced atherosclerosis from intractably high levels of cholesterol and LDL. Clearly, new therapeutic strategies are needed. Accordingly, apoB, a protein not amenable to inhibition by conventional small-molecule- or protein-based therapeutics, was selected as a potential clinical target for development of siRNA therapeutics.
Using conventional bioinformatics, 84 siRNAs specific for both human and mouse apoB mRNA were designed and synthesized (data not shown). These apoB-siRNAs were screened for their ability to reduce apoB mRNA and protein levels, as measured by polymerase chain reaction with reverse transcription (RT–PCR) and enzyme-linked immunosorbent assay (ELISA), respectively, in HepG2 liver cells after transfection at a concentration of 100 nM. Five apoB-siRNAs were identified that reduced both mRNA and protein levels by >70%. Because exonucleolytic degradation is the predominant mechanism for siRNA degradation in serum, two selected apoB-siRNAs (apoB-1-siRNA and apoB-2-siRNA) and one four-nucleotide mismatch control for apoB-1-siRNA (mismatch-siRNA) were stabilized at the 3′ end of the sense and antisense strands by phosphorothioate backbone modifications and additional incorporation of two 2’-O-methyl nucleotides at the 3′ end of the antisense strand. Chol-siRNAs were synthesized by linkage of cholesterol to the 3′ end of the sense strand via a pyrrolidine linker. Chol-apoB-1-siRNA was significantly more stable than unconjugated apoB-1-siRNA in human serum: gel electrophoresis showed >50% intact chol-apoB-1-siRNA after a 1 h incubation at 37 °C compared with <5% intact unconjugated apoB-1-siRNA. Similar data were obtained for chol-apoB-2-siRNA, although this siRNA was less stable than chol-apoB-1-siRNA. Dose response curves for the activity of conjugated and unconjugated apoB-specific and control siRNAs were measured in HepG2 cells using transfection. Two conjugated control siRNAs (chol-luc-siRNA and chol-mismatch-siRNA) showed no significant inhibition of apoB protein expression at concentrations as high as 30 nM. In contrast, three specific siRNAs (unconjugated apoB-1-siRNA, chol-apoB-1-siRNA and chol-apoB-2-siRNA) showed dose-dependent silencing of apoB protein expression based on apoB ELISA measurements—IC50 values of 0.5 nM, 5 nM and 8 nM were calculated, respectively.
In vivo studies with modified siRNAs
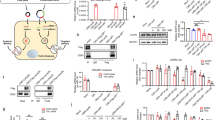
To demonstrate the ability of chol-apoB-siRNAs to silence apoB expression in vivo, experiments were first performed in C57BL/6 mice fed a normal chow diet. siRNAs were administered by tail-vein injection with normal volume (0.2 ml) and normal pressure. Biodistribution of siRNAs was assessed by RPA of siRNAs in tissue samples from liver and jejunum obtained 24 h after the last injection. Significant levels of chol-luc-siRNA, chol-apoB-1-siRNA and chol-mismatch-siRNA were detected in liver and jejunum (100–200 ng g-1 tissue for chol-apoB-1-siRNA), whereas levels of unconjugated apoB-1-siRNA were below our detection limit (Fig. 1). Levels of chol-apoB-2-siRNA were also detected but at levels approximately 10% of those observed for other chol-siRNAs.
An RPA was used to detect siRNAs in pooled liver and jejunum tissue lysates from animals injected with saline (- ), chol-luc-siRNA (A), chol-mismatch-siRNA (B), unconjugated apoB-1-siRNA (C2) or chol-apoB-1-siRNA (C1). Detection by RPA of endogenous miRNAs in liver (miR-122) and jejunum (miR-143) served as an internal loading control.
The primary measure of RNAi-mediated effects is the reduction (that is, silencing) of the target mRNA. To measure silencing of apoB mRNA, we used a branched-DNA (bDNA) detection method and bDNA probes (Fig. 2a) to quantify apoB mRNA levels in liver and jejunum, two organs where apoB is known to be expressed. As shown in Fig. 2b, mice treated with chol-apoB-1-siRNA and chol-apoB-2-siRNA showed statistically significant reductions (mean ± s.d.; 57 ± 6% and 36 ± 8%, respectively) in apoB mRNA levels in liver samples as compared with saline control (P < 0.0001). In jejunum tissue samples, mice injected with chol-apoB-1-siRNA and chol-apoB-2-siRNA showed an even more substantial reduction in apoB mRNA levels of 73 ± 10% and 51 ± 13%, respectively, as compared with saline control (P < 0.0001). Individual animal results for apoB mRNA reduction in the liver are shown in Fig. 2c and demonstrate the consistent and robust effect observed for specific chol-siRNAs as compared with other treatment groups. Similar results were observed for apoB mRNA reduction in the jejunum from individual animals (data not shown). Owing to the extended length of the apoB mRNA, two additional probes at the distal ends of the apoB open reading frame (ORF) were designed. As measured with the three divergent probe sets, identical levels of apoB mRNA reduction were detected for animals treated with chol-apoB-1-siRNA and chol-apoB-2-siRNA (Fig. 2d). These data suggest a uniform and rapid degradation of apoB mRNA after treatment with chol-apoB-siRNAs, and argue against the potential existence of truncated amino-terminal apoB protein fragments translated from incompletely degraded siRNA-cleavage products, as has been reported for ribozyme-mediated cleavage of apoB mRNA12.
Treatment groups comprised saline control (n = 10), chol-luc-siRNA control (n = 10), chol-mismatch-siRNA control (n = 10), unconjugated apoB-1-siRNA (n = 10), chol-apoB-1-siRNA (n = 10) and chol-apoB-2-siRNA (n = 7). bDNA measurements were performed with probe set II. Error bars represent the standard deviation (s.d.) of the mean. Statistical analysis was by analysis of variance (ANOVA) with Bonferroni post-hoc t-test, one-tailed. Asterisk, P < 0.0001 compared with saline control animals. a, Schematic representation of the apoB mRNA illustrating the binding regions of three bDNA probe sets in relation to the two siRNA cleavage sites. b, Effects of siRNA administration on mean apoB mRNA levels. c, apoB mRNA levels from individual mice treated with saline or siRNAs. Data are mean values from three liver samples from each individual animal. d, Effects of siRNA administration on the reduction of apoB mRNA measured by bDNA assays using three different probe sets.
Silencing of the apoB mRNA would be expected to result in a corresponding reduction in apoB protein levels. An ELISA-based method specific for detection of apoB-100 protein was used to measure the effects of chol-apoB-siRNA treatment on plasma levels of apoB protein. In addition to the effects on apoB mRNA levels, treatment with chol-apoB-1-siRNA and chol-apoB-2-siRNA reduced plasma levels of apoB-100 protein 24 h after siRNA treatment by 68 ± 14% and 31 ± 18%, respectively, compared with levels in saline-treated control animals (Fig. 3). These results achieved statistical significance (P < 0.0001) for the group treated with the more potent and stable chol-apoB-1-siRNA. As the LF3 antibody used in this study recognizes only apoB-100, and not apoB-48, the observed apoB-100 reduction may underestimate the full effect of chol-apoB-1-siRNA at the protein level.
To confirm the physiological relevance of apoB mRNA silencing on lipoprotein metabolism, we characterized the effect of siRNA treatment and the resulting reduction of apoB protein levels on lipoprotein profiles and cholesterol levels. Using an NMR-based method, complete lipoprotein profiles were generated and concentrations of chylomicrons, very-low-density lipoprotein (VLDL), LDL and high-density lipoprotein (HDL) particles were calculated (Fig. 4a). As expected, HDL represented the predominant lipoprotein fraction in mouse plasma. Similar to results observed in heterozygous knockout mice for apoB9, treatment with chol-apoB-1-siRNA resulted in a 25% reduction in HDL particle concentration (Fig. 4b). Furthermore, treatment of mice with chol-apoB-1 siRNA resulted in an almost 50% reduction of chylomicron levels and an approximately 40% reduction in LDL levels, whereas VLDL levels were not altered. Treatment with either of the control siRNAs did not change the lipoprotein profile significantly. In addition to reductions in lipoprotein concentrations, in vivo silencing of apoB by chol-apoB-1-siRNA led to a significant reduction (37 ± 11%; P < 0.0001) of total plasma cholesterol as compared with saline control animals (Fig. 4c). Treatment with the less potent chol-apoB-2-siRNA failed to show significant reductions in cholesterol, consistent with the reduced activity of this chol-siRNA on apoB mRNA and protein levels. Treatment with chol-apoB-1-siRNA also resulted in a 44% decrease in LDL-associated cholesterol, consistent with the effects observed on apoB protein levels. In aggregate, the effects on cholesterol reduction and lipoprotein profiles would be considered highly clinically significant in patients with hypercholesterolaemia, and actually exceed the level of cholesterol reduction observed in heterozygous apoB knockout mice9.
a, Lipoprotein profile of pooled plasma samples from treatment groups determined by NMR analysis. b, Relative reduction of lipoprotein classes for the siRNA treatment groups normalized against the average levels of saline control group. c, Effects of siRNA administration on plasma cholesterol and LDL cholesterol. Plasma cholesterol was determined by enzymatic assay and LDL cholesterol calculated from NMR measurements. Error bars represent the s.d. of the mean. Statistical analysis was by ANOVA with Bonferroni post-hoc t-test, one-tailed. Asterisk, P < 0.0001 compared with saline and chol-mismatch-siRNA control animals. NMR data are based on single measurements of pooled plasma from treatment groups.
To extend our findings of in vivo silencing by chol-apoB-siRNAs in normal mice, we performed an additional study in a human apoB transgenic mouse model13. These mice express human apoB-100 in liver and have elevated levels of apoB as compared with normal mice; when fed a high-fat diet, these mice develop severe atherosclerosis14. In our experiments, we administered saline, chol-mismatch-siRNA and chol-apoB-1-siRNA to apoB transgenic mice fed a normal chow diet. As shown in Fig. 5, chol-apoB-1-siRNA brought about a significant reduction of endogenous murine apoB expressed in both liver and jejunum tissue samples (P < 0.0001, relative to saline and chol-mismatch-siRNA treatment). Relative to the saline control, levels of murine apoB mRNA were reduced by 57 ± 10% in liver and 42 ± 12% in jejunum. In addition, chol-apoB-1-siRNA, which was selected in part owing to its sequence identity to both human and mouse apoB, showed significant silencing of the human transgene expressed in the liver, where human apoB mRNA was silenced by 60 ± 10% (P < 0.0001). In contrast to these effects, chol-mismatch-siRNA showed no effect on mouse or human apoB mRNA levels. These results confirm the effect of specific chol-siRNAs on apoB silencing in a different mouse model. Moreover, this specific chol-siRNA was shown to silence a transgenic human mRNA in vivo.
Reduction of human and mouse apoB mRNA levels in mice transgenic for human apoB that received saline (n = 8), chol-mismatch-siRNA (n = 8) and chol-apoB-1-siRNA (n = 8). Statistical analysis was by ANOVA with Bonferroni post-hoc t-test, one-tailed. Asterisk, P < 0.0001 compared with saline and chol-mismatch-siRNA control animals. Error bars illustrate s.d. of the mean.
An important consideration for siRNA-mediated inhibition of gene expression is whether the observed effects are specific and not due to nonspecific “off target” effects15 and potential interferon responses16, which have been reported with siRNAs in vitro and other oligonucleotide-based approaches in vivo. In our experiments, the effects of apoB-specific, cholesterol-conjugated siRNAs were seen with two divergent siRNAs targeting separate sequence regions of the apoB mRNA. Furthermore, the in vivo silencing of apoB by these siRNAs was specific as neither an irrelevant siRNA (chol-luc-siRNA) nor a mismatch control siRNA (chol-mismatch-siRNA)—although present at comparable concentrations in liver and jejunum—mediated a significant reduction in apoB mRNA, plasma apoB protein levels, or total cholesterol. Finally, the silencing of apoB mRNA by chol-apoB-siRNAs in liver as measured by bDNA assay and normalization to GAPDH mRNA was also demonstrated with normalization to three other liver mRNAs, including factor VII, glucose-6-phosphatase and VEGF (Supplementary Fig. 1).
Determination of in vivo mechanism of action
To prove that the in vivo activity was due to siRNA-directed cleavage, we characterized specific mRNA cleavage products using a modified 5′-RACE (rapid amplification of cDNA ends) technique previously used to demonstrate microRNA (miRNA)-directed mRNA cleavage in plants17 and mouse embryos18. As it relates to the specific cleavage of apoB mRNA by apoB-1-siRNAs, total RNA from mice in the different treatment groups was isolated, and then PCR was used to reveal fragments of the predicted length in animals receiving chol-apoB-1-siRNA treatment (Fig. 6a). Identity of the PCR products was confirmed by direct sequencing of the excised bands, which demonstrated that cleavage occurred at the predicted position for the siRNA duplex. Indeed, sequencing revealed cleavage after position 10,061 of the apoB ORF, exactly ten nucleotides downstream of the 5′ end of the siRNA antisense strand. Specific cleavage fragments were detected in both liver and jejunum of animals receiving chol-apoB-1-siRNA treatment (Fig. 6b). No fragments were detected in tissues of animals receiving control siRNAs (chol-luc-siRNA or chol-mismatch-siRNA) or saline. As expected, in this 5′-RACE experiment of apoB mRNA cleavage mediated by chol-apoB-1-siRNA, no fragments were detected in tissues from animals receiving the alternative apoB-specific siRNA (chol-apoB-2-siRNA). Notably, a low level of specific cleavage product was detected in the jejunum of animals receiving the unconjugated apoB-1-siRNA despite no evidence for significant knockdown of total apoB mRNA levels by this siRNA. This indicates that some unconjugated apoB-1-siRNA is able to enter epithelial cells of the jejunum after systemic administration despite lacking cholesterol conjugation. Together, these data demonstrate that inhibition of apoB was achieved by an RNAi mechanism of action. To our knowledge, this is the first demonstration of silencing of an endogenous gene in mammals by a mechanism of RNAi-mediated degradation of the target mRNA.
. a, Schematic representation of the apoB mRNA illustrating siRNA cleavage sites and RACE strategy to detect cleavage product. Cleaved mRNA ligated to an RNA adaptor was reverse transcribed using primer GSP. b, Agarose gel of 5′-RACE–PCR amplification, using the primer pair GR5′ and Rev2, showing specific cleavage products in liver and jejunum. Treatment groups are: A, saline; B, chol-luc-siRNA; C, chol-mismatch-siRNA; D, apoB-1-siRNA; E, chol-apoB-1-siRNA; F, chol-apoB-2-siRNA.
Discussion
Our findings demonstrate that RNAi can be used to silence endogenous genes involved in the cause or pathway of human disease with a clinically acceptable formulation and route of administration by means of systemic delivery. In our study, we have shown that the mechanism of action for chemically modified siRNAs was by RNAi-mediated degradation of the target mRNA. Chol-apoB-siRNAs, but not unconjugated apoB-siRNAs, showed biological activity, demonstrating an important role for cholesterol conjugation of siRNAs to achieve systemic in vivo activity, and suggesting the opportunity to further optimize systemic activity through chemical conjugation strategies. Indeed, further optimization is warranted to achieve improved in vivo potency for chol-siRNAs at doses and dose regimens that are clinically acceptable. Nevertheless, these findings hold promise for the development of a new class of therapeutics that harnesses the RNAi mechanism. Of particular interest is the use of RNAi therapeutics to silence genes (such as the apoB gene) or mutated or variant alleles whose proteins are refractory to the discovery of traditional small molecules or biotherapeutic drugs.
Methods
Synthesis of siRNAs
The siRNAs used in this study consisted of a 21-nucleotide sense strand and a 23-nucleotide antisense strand resulting in a two-nucleotide overhang at the 3′ end of the antisense strand. apoB-1-siRNA (ORF position 10049–10071): sense 5′-GUCAUCACACUGAAUACCAA*U-3′, antisense 5′-AUUGGUAUUCAGUGUGAUGAc*a*C-3′; chol-apoB-1-siRNA: sense 5′-GUCAUCACACUGAAUACCAAU*chol-3′, antisense 5′-AUUGGUAUUCAGUGUGAUGAc*a*C-3′; chol-mismatch-siRNA: sense 5′-GUGAUCAGACUCAAUACGAAU*chol-3′, antisense 5′-AUUCGUAUUGAGUCUGAUCAc*a*C-3′; chol-apoB-2-siRNA (ORF position 327–349): sense 5′-AGGUGUAUGGCUUCAACCCUG*chol-3′, antisense 5′-CAGGGUUGAAGCCAUACACCu*c*U-3′; chol-luc-siRNA: sense 5′-GAACUGUGUGUGAGAGGUCCU*chol-3′, antisense 5′-AGGACCUCUCACACACAGUUc*g*C-3′. The lower-case letters represent 2’-O-methyl-modified nucleotides; asterisks represent phosphorothioate linkages.
RNA oligonucleotides were synthesized using commercially available 5′-O-(4,4′-dimethoxytrityl)-3′-O-(2-cyanoethyl-N,N-diisopropyl) phosphoramidite monomers of uridine (U), 4-N-benzoylcytidine (CBz), 6-N-benzoyladenosine (ABz) and 2-N-isobutyrylguanosine (GiBu) with 2′-O-t-butyldimethylsilyl protected phosphoramidites and the corresponding 2′-O-methyl phosphoramidites according to standard solid phase oligonucleotide synthesis protocols19. After cleavage and de-protection, RNA oligonucleotides were purified by anion-exchange high-performance liquid chromatography and characterized by ES mass spectrometry and capillary gel electrophoresis. RNA with phosphorothioate backbone at a given position was achieved by oxidation of phosphite with Beaucage reagent20 during oligonucleotide synthesis. Chol-siRNAs were synthesized using the same protocols as above except that the RNA synthesis started from a controlled-pore glass solid support carrying a cholesterol-aminocaproic acid-pyrrolidine linker (V.Ke., K.G.R. and M.M., unpublished data). For this support, the first nucleotide linkage was achieved using a phosphorothioate linkage to provide additional 3′-exonuclease stability. To generate siRNAs from RNA single strands, equimolar amounts of complementary sense and antisense strands were mixed and annealed, and siRNAs were further characterized by native gel electrophoresis.
In vitro activity and stability assays
To determine in vitro activity of siRNAs, HepG2 cells were transfected with siRNAs using oligofectamine (Invitrogen) and siRNA concentrations ranging from 0.1, 0.3, 1, 3, 10 to 30 nM. apoB protein concentration was determined from cell culture supernatant by a sandwich ELISA capturing apoB with a polyclonal goat anti-human apoB antibody (Chemicon International). apoB detection was performed with a horseradish peroxidase-conjugated goat anti-human apoB-100 polyclonal antibody (Academy Bio-Medical Company). The remaining apoB protein content was calculated as the ratio of apoB protein concentration in the supernatant of cells treated with the apoB-specific siRNA duplex to the apoB concentration in the supernatant of cells treated with an unspecific control siRNA duplex. Mouse serum (Sigma-Aldrich Chemie GmbH) was used for stability assays. Double-stranded RNAs (5 µM) were incubated in 95% serum, and the mixture was incubated at 37 °C for various lengths of time (for example, 0, 15 or 30 min, or 1, 2, 4, 8, 16 or 24 h). siRNAs were isolated by hot phenol extraction in the presence of sodium dodecyl sulphate followed by ethanol precipitation. Re-suspended RNA samples were run on a denaturing 14% polyacrylamide gel containing 20% formamide for 2 h at 45 mA. RNA bands were visualized by staining with the ‘Stains-All’ reagent (Sigma-Aldrich Chemie GmbH) according to the manufacturer's instructions.
In vivo silencing experiments
C57BL/6 mice received, on three consecutive days, tail vein injections of saline or different siRNAs. All siRNAs were administered at doses of 50 mg kg-1 in approximately 0.2 ml per injection. Measurements of apoB mRNA, apoB protein levels, lipoprotein concentrations and plasma cholesterol content were performed 24 h after the last i.v. injection. Experiments were carried out in a blinded fashion. The same experimental design was used for experiments with the human apoB transgenic mice (1004-T hemizygotes, Taconic).
In vivo bioanalytical methods
An RPA, using radiolabelled probes complementary to the antisense strands, was used to detect siRNAs in pooled liver and jejunum tissue lysates from animals treated with saline or siRNAs. RPA for endogenous miRNAs was used as a loading control for jejunum (miR-143, sequence 5′-UGAGAUGAAGCACUGUAGCUCA-3′) and liver (miR-122, 5′-UGGAGUGUGACAAUGGUGUUUG-3′).
The QuantiGene assay (Genospectra) was used to quantify the reduction of mouse apoB mRNA in liver and jejunum tissue after siRNA treatment. Small uniform tissue samples where collected 24 h after the last injection. Lysates from three tissue samples per animal were directly used for apoB and GAPDH mRNA quantification, and the ratio of apoB and GAPDH mRNA was calculated and expressed as a group average relative to the saline control group. Specific probes for detection of apoB mRNA levels were designed to the following regions of the apoB mRNA ORF: probe set I 83–385; probe set II 5,045–5,673; probe set III 12,004–12,411. Furthermore, apoB mRNA reduction in liver was quantified from purified (RNeasy mRNA isolation kit, Qiagen), pooled mRNA for each treatment group. As well as GAPDH, factor VII, glucose-6-phosphatase and VEGF mRNAs were also used for normalization.
ELISA was used to quantify the reduction of apoB-100 protein levels in mouse plasma after siRNA treatment. apoB-100 from plasma samples of individual animals was detected using the primary antibody LF3 against mouse apoB-100 (gift of S. Young; see ref. 21). Levels were normalized to plasma volume and expressed as group averages relative to the saline control group.
Total cholesterol levels in the plasma were measured using the Cholesterol detection kit (Diasys). For NMR determination of the plasma lipoprotein profile a Bruker DRX 600 with cryoprobe head was used (LipoFIT Analytic GmbH). Single measurements of 500 µl mouse plasma (pooled from ten animals per treatment group) were performed. The lipoprotein subclass distribution was calculated from the NMR data by using computer algorithms that are based on human blood standards22. The particle number for lipoprotein classes was calculated based on the correlation of known particle size and composition with the experimentally determined NMR signal intensity. On the basis of this correlation, the cholesterol content in the LDL fraction was computed. The cholesterol values calculated from NMR data were confirmed by the presence of comparable levels of total cholesterol in plasma and HDL-cholesterol as determined by enzymatic assays.
5′-RACE analysis
Total RNA (5 µg) from pooled liver and jejunum samples from animals treated with different siRNAs was ligated to a GeneRacer adaptor (Invitrogen) without prior treatment. Ligated RNA was reverse transcribed using a gene-specific primer (GSP: 5′-CTCCTGTTGCAGTAGAGTGCAGCT-3′). To detect cleavage products, PCR was performed using primers complementary to the RNA adaptor (GR5’: 5′-CTCTAGAGCGACTGGAGCACGAGGACACTA-3′) and apoB mRNA (Rev2: 5′-ACGCGTCGACGTGGGAGCATGGAGGTTGGCAGTTGTTC-3′). Amplification products were resolved by agarose gel electrophoresis and visualized by ethidium bromide staining. The identity of specific PCR products was confirmed by sequencing of the excised bands.
References
Novina, C. D. & Sharp, P. A. The RNAi revolution. Nature 430, 161–164 (2004)
Scherr, M., Battmer, K., Dallmann, I., Ganser, A. & Eder, M. Inhibition of GM-CSF receptor function by stable RNA interference in a NOD/SCID mouse hematopoietic stem cell transplantation model. Oligonucleotides 13, 353–363 (2003)
Song, E. et al. RNA interference targeting Fas protects mice from fulminant hepatitis. Nature Med. 9, 347–351 (2003)
Reich, S. J. et al. Small interfering RNA (siRNA) targeting VEGF effectively inhibits ocular neovascularization in a mouse model. Mol. Vis. 9, 210–216 (2003)
Zhang, X. et al. Small interfering RNA targeting heme oxygenase-1 enhances ischemia-reperfusion-induced lung apoptosis. J. Biol. Chem. 279, 10677–10684 (2004)
Dorn, G. et al. siRNA relieves chronic neuropathic pain. Nucleic Acids Res. 32, e49 (2004)
Lorenz, C., Hadwiger, P., John, M., Vornlocher, H.-P. & Unverzagt, C. Steroid and lipid conjugates of siRNAs to enhance cellular uptake and gene silencing in liver cells. Bioorg. Med. Chem. Lett. 14, 4975–4977 (2004)
Burnett, J. R. & Barrett, P. H. Apolipoprotein B metabolism: tracer kinetics, models, and metabolic studies. Crit. Rev. Clin. Lab. Sci. 39, 89–137 (2002)
Farese, R. V. Jr, Ruland, S. L., Flynn, L. M., Stokowski, R. P. & Young, S. G. Knockout of the mouse apolipoprotein B gene results in embryonic lethality in homozygotes and protection against diet-induced hypercholesterolemia in heterozygotes. Proc. Natl Acad. Sci. USA 92, 1774–1778 (1995)
Brown, M. S. & Goldstein, J. L. A receptor-mediated pathway for cholesterol homeostasis. Science 232, 34–47 (1986)
Glueck, C. J. et al. Prospective 10-year evaluation of hypobetalipoproteinemia in a cohort of 772 firefighters and cross-sectional evaluation of hypocholesterolemia in 1,479 men in the National Health and Nutrition Examination Survey I. Metabolism 46, 625–633 (1997)
Enjoji, M., Wang, F., Nakamuta, M., Chan, L. & Teng, B. B. Hammerhead ribozyme as a therapeutic agent for hyperlipidemia: production of truncated apolipoprotein B and hypolipidemic effects in a dyslipidemia murine model. Hum. Gene Ther. 11, 2415–2430 (2000)
Linton, M. F. et al. Transgenic mice expressing high plasma concentrations of human apolipoprotein B100 and lipoprotein(a). J. Clin. Invest. 92, 3029–3037 (1993)
Purcell-Huynh, D. A. et al. Transgenic mice expressing high levels of human apolipoprotein B develop severe atherosclerotic lesions in response to a high-fat diet. J. Clin. Invest. 95, 2246–2257 (1995)
Jackson, A. L. et al. Expression profiling reveals off-target gene regulation by RNAi. Nature Biotechnol. 21, 635–637 (2003)
Bridge, A. J., Pebernard, S., Ducraux, A., Nicoulaz, A.-L. & Iggo, R. Induction of an interferon response by RNAi vectors in mammalian cells. Nature Genet. 34, 263–264 (2003)
Llave, C., Xie, Z., Kasschau, K. D. & Carrington, J. C. Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science 20, 2053–2056 (2002)
Yekta, S., Shih, I. H. & Bartel, D. P. MicroRNA-directed cleavage of HOXB8 mRNA. Science 304, 594–596 (2004)
Damha, M. J. & Ogilvie, K. K. Oligoribonucleotide synthesis. The silyl-phosphoramidite method. Methods Mol. Biol. 20, 81–114 (1993)
Iyer, R. P., Egan, W., Regan, J. B. & Beaucage, S. L. 3H–1,2-benzodithiole-3-one 1,1-dioxide as an improved sulfurizing reagent in the solid phase synthesis of oligodeoxyribonucleoside phosphorothioates. J. Am. Chem. Soc. 112, 1253–1254 (1990)
Zlot, C. H. et al. Generation of monoclonal antibodies specific for mouse apolipoprotein B-100 in apolipoprotein B-48-only mice. J. Lipid Res. 40, 76–84 (1999)
Hammad, S. M. et al. Lipoprotein subclass profiles of hyperlipidemic diabetic mice measured by nuclear magnetic resonance spectroscopy. Metabolism 52, 916–921 (2003)
Acknowledgements
We thank P. Sharp for his advice and creative input. We are grateful to J. Maraganore and T. Ulich for their support and encouragement. We would like to thank S. Young for the LF3 anti-mouse apoB antibody; D. Bartel and S. Yekta for advice on the 5′-RACE assay; S. Young and M. Stoffel for valuable discussions; and LipoFIT Analytic GmbH and the Institute for Biophysics and Physical Biochemistry of the University of Regensburg for the characterization of lipoprotein particles by NMR. For technical assistance we thank P. Deuerling, F. Hertel, S. Leuschner, N. Linke, A. Müller, G. Ott, H. Schübel, S. Shanmugam, M. Duckman and C. Auger.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
All authors are employees of Alnylam.
Supplementary information
Supplemental Figure 1
Specificity of apoB mRNA reduction in wild-type mice. siRNA-mediated reduction of apoB mRNA in liver mRNA pools from wild-type mice that received saline (n=10), Chol-mismatch-siRNA (n=10), and Chol-apoB-1-siRNA (n=10). The apoB mRNA was normalized to 4 reference mRNAs: glyceraldehyde 3-phosphate dehydrogenase (GAPDH), glucose-6-phosphatase (G-6-P), factor VII (FVII) and vascular endothelial growth factor (VEGF). Data represent the mean of three independent measurements. Error bars illustrate the standard deviation of the mean. (JPG 18 kb)
Rights and permissions
About this article
Cite this article
Soutschek, J., Akinc, A., Bramlage, B. et al. Therapeutic silencing of an endogenous gene by systemic administration of modified siRNAs. Nature 432, 173–178 (2004). https://doi.org/10.1038/nature03121
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nature03121
This article is cited by
-
RNA interference in the era of nucleic acid therapeutics
Nature Biotechnology (2024)
-
Oligonucleotide therapeutics and their chemical modification strategies for clinical applications
Journal of Pharmaceutical Investigation (2024)
-
Biomembrane-derived nanoplexes for SiRNAs-pioneer innovation in delivery to lung adenocarcinoma
Journal of Nanoparticle Research (2024)
-
Structural optimization of siRNA conjugates for albumin binding achieves effective MCL1-directed cancer therapy
Nature Communications (2024)
-
In vivo PAR-CLIP (viP-CLIP) of liver TIAL1 unveils targets regulating cholesterol synthesis and secretion
Nature Communications (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.