Abstract
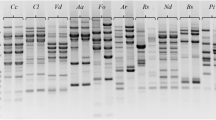
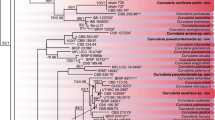
Isolates of three closely related pea pathogens,Ascochyta pisi, Ascochyta pinodes (teleomorphMycosphaerella pinodes) andPhoma medicaginis var. pinodella, were compared by means of isozyme analysis and restriction fragment length polymorphisms (RFLP) of amplified rDNA spacers. Three enzyme systems differentiatedA. pisi fromM. pinodes andP. m. pinodella. The internal transcribed spacers (ITSs) of the three fungi showed no intraspecific and very little interspecific variation after digestion with 12 endonucleases. Digestion of the intergenic spacer (IGS) withHinfI, andSau3A revealed uniformity inA. pisi patterns which consistently differed from those ofM. pinodes andP. m. pinodella. No clear distinction could be made between the latter two fungi which both showed intraspecific variability. Both biochemical and molecular markers thus discriminated between twoAscochyta species. The results also indicated a closer relationship between two organisms belonging to different genera (Ascochyta andPhoma) than between two species of the same genus (Ascochyta).
Similar content being viewed by others
References
Boerema GH, Bollen GJ (1975) Conidiogenesis and conidial septation as differentiating criteria betweenPhoma andAscochyta. Persoonia 8:111–144
Bonde MR, Peterson GL, Emmett RW, Menge JA (1991) Isozyme comparison ofSeptoria isolates associated with citrus in Australia and the United States. Phytopathology 81:517–521
Chandon JL, Pinson S (1981) In: Masson (ed) Analyses typologiques. Théories et applications
Damaj M, Jabaji-Hare SH, Charest P-M (1993) Isozyme variation and genetic relatedness in binucleateRhizoctonia species. Phytopathology 83:864–871
Dellaporta SL, Wood J, Hicks JB (1983) A plant DNA mini-preparation: 2nd edn. Plant Mol Biol Rep 1:19–21
Erland S, Herion B, Martin F, Glover LA, Alexander IJ (1994) Identification of the ectomycorrhizal basidiomyceteTylospora fibrillosa Donk. by RFLP of the PCR-amplified ITS and IGS regions of ribosomal DNA. New Phytol 126:525–532
Gall C, Balesdent M-H, Desthieux I, Robin P, Rouxel T (1995) Polymorphism of Tox°Leptosphaeria maculans isolates as revealed by soluble protein and isozyme electrophoresis. Mycol Res 99:221–229
Garber RC, Turgeon BG, Selker EU, Yoder OC (1988) Organization of ribosomal RNA genes in the fungusCochhobolus heterostrophus. Curr Genet 14:573–582
Hellman R, Christ BJ (1991) Isozyme variation of physiologic races ofUstilago hordei. Phytopathology 81:1536–1540
Hwang SF, Lopetinsky K, Evans IR (1991) Effects of seed infection byAscochyta spp., fungicide treatment, and cultivar on yield parameters of field pea under field conditions. Can Plant Dis Survey 71:169–172
Jones LK (1927) Studies of the nature and control of blight, leaf and pod spot and foot rot of peas caused by species ofAscochyta. NY State Agric Exp Stn Bull 547:1–45
Julian AM, Lucas JA (1990) Isozyme polymorphism in pathotypes ofPseudocercosporella herpotrichoides and related species from cereals. Plant Pathol 39:178–190
Kistler HC, Miao VPW (1992) New modes of genetic change in filamentous fungi. Annu Rev Phytopathol 30:131–152
Klassen GR, Buchko J (1990) Subrepeat structure of the intergenic region in the ribosomal DNA of the oomycetous fungusPythium ultimum. Curr Genet 17:125–127
Lee SB, White TJ, Taylor JW (1993) Detection ofPhytophthora species by oligonucleotide hybridization to amplified ribosomal DNA spacers. Phytopathology 83:177–181
Lévesque AC, Vrain TC, DeBoer SH (1994) Production of species-specific probes for differentPythium species using PCR and ribosomal DNA. Phytopathology 84:474–478
Louanchi M (1993) Contribution à l'étude deRigidoporus lignosus agent du pourridie blanc des racines d'Hevea brasiliensis: étude de la diversité génétique des populations et détection par les méthodes immunoenzymatiques. PhD thesis. Université Paris XI. Orsay
Maniatis T, Fritsch EF, Sambrook J (1982) Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, New York
Mathur RS, Barnett HL, Lilly VG (1950) Sporulation ofColletotrichum lindemuthianum in culture. Phytopathology 40:104–114
Meijer G, Megnegneau B, Linders EGA (1994) Variability for isozyme, vegetative compatibility and RAPD markers in natural populations ofPhomopsis subordinaria. Mycol Res 98:267–276
Montel E, Bridge PD, Sutton BC (1991) An integrated approach toPhoma systematics. Mycopathologia 115:89–103
Nazar RN, Hu X, Schmidt J, Culham D, Robb J (1991) Potential use of PCR-amplified ribosomal intergenic sequences in the detection and differentiation ofVerticillium wilt pathogens. Physiol Mol Plant Pathol 39:1–11
Nooderloos ME, De Gruyter J, Van Eijk GW, Roeijmans HJ (1993) Production of dendritic crystals in pure cultures ofPhoma andAscochyta and its value as a taxonomic character relative to morphology, pathology and cultural characteristics. Mycol Res 97:1343–1350
O'Donnell K (1992) Ribosomal DNA internal transcribed spacers are highly divergent in the phytopathogenic ascomyceteFusarium sambucinum (Gibberella pulicaris). Curr Genet 22:213–220
Petrunak DM, Christ BJ (1992) Isozyme variability inAlternaria solani andA. alternata. Phytopathology 82:1343–1347
Poupard P, Simonet P, Cavelier N, Bardin R (1993) Molecular characterization ofPseudocercosporella herpotrichoides isolates by amplification of ribosomal DNA internal spacers. Plant Pathol 42:873–881
Sherriff C, Whelan MJ, Arnold GM, Bailey JA (1995) rDNA sequence analysis confirms the distinction betweenColletotrichum graminicola andC. sublineolum. Mycol Res 99:475–478
Sneath PHA, Sokal RR (1973) Numerical taxonomy. W.H. Freeman and Co., San Francisco, USA
Tisserat NA, Hulbert SH, Sauer KM (1994) Selective amplification of rDNA internal transcribed spacer regions to detectOphiosphaerella korrae andO. herpotricha. Phytopathology 84:478–482
Wallen VR (1965) Field evaluation and the importance of theAscochyta complex on peas. Can J Plant Sci 45:27–33
White JF, Morgan-Jones G (1987) Studies in the genus Phoma. VI. ConcerningPhoma medicaginis var.pinodella. Mycotaxon 28:241–248
White TJ, Bruns T, Lee S, Taylor J (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ (eds) PCR protocols, a guide to methods and applications. Academic Press, San Diego, CA, USA, pp 315–322
Author information
Authors and Affiliations
Additional information
Communicated by P.J.G.M. de Wit
Rights and permissions
About this article
Cite this article
Faris-Mokaiesh, S., Boccara, M., Denis, J.B. et al. Differentiation of the “Ascochyta complex” fungi of pea by biochemical and molecular markers. Curr Genet 29, 182–190 (1996). https://doi.org/10.1007/BF02221583
Received:
Issue Date:
DOI: https://doi.org/10.1007/BF02221583