Abstract
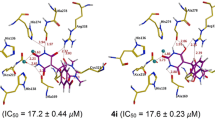
In the current study, amphiphilic peptides were designed and screened against Jack bean urease by using computer aided drug discovery approach. The result showed that out of thirty-eight amphiphilic peptides 1, 3, 12, 18, 30, and 33 exhibit stronger binding affinity with the active site of the enzyme through chelation of charged amino acids with the nickel ions i.e., Ni+2 841 and Ni+2 842 as well as hydrophobic contacts of the nonpolar tail with the nonpolar residues in the active site. The selected amphiphilic peptides were synthesized by solid-phase peptide synthesis strategy, characterized by fast atomic bombardment mass spectroscopy (FAB-MS) and nuclear magnetic resonance spectroscopy (1H and 13C-NMR) and in vitro urease inhibitory activity of amphiphilic peptides was studied. Amphiphilic peptides 12 and 33 showed excellent urease inhibitory activity, (p < 0.001) with IC50 values 20.5 ± 0.01, and 28.1 ± 0.03 µM respectively, which was considerably better than thiourea used as positive control.

Highlights
-
Molecular docking.
-
Solid-phase synthesis of amphiphilic peptides.
-
FAB MS-MS and 1H and 13C NMR study of amphiphilic peptides.
-
Urease inhibitory activity.





Similar content being viewed by others
References
Redgrave LS, Sutton SB, Webber MA, Piddock LJ. Fluoroquinolone resistance: mechanisms, impact on bacteria, and role in evolutionary success. Trends Microbiol. 2014;22:438–45. https://doi.org/10.1016/j.tim.2014.04.007.
Mobley HL, Hausinger RP. Microbial ureases: significance, regulation, and molecular characterization. Microbiol Mol Biol Rev. 1989;53:85–108. https://mmbr.asm.org/content/53/1/85 https://mmbr.asm.org/content/53/1/85.
Karplus PA, Pearson MA, Hausinger RP. 70 years of crystalline urease: what have we learned? Acc Chem. 1997;30:330–7. https://doi.org/10.1021/ar960022j.
Collins CM, D’Orazio SE. Bacterial ureases: structure, regulation of expression and role in pathogenesis. Mol Microbiol. 1993;9:907–13. https://doi.org/10.1111/j.1365-2958.1993.tb01220.x.
Burne RA, Chen YY. Bacterial ureases in infectious diseases. Microbes Infect. 2000;2:533–42. https://doi.org/10.1016/S1286-4579(00)00312-9.
Benini S, Rypniewski WR, Wilson KS, Miletti S, Ciurli S, Mangani S. A new proposal for urease mechanism based on the crystal structures of the native and inhibited enzyme from Bacillus pasteurii: why urea hydrolysis costs two nickels. Structure. 1999;7:205–16. https://doi.org/10.1016/S0969-2126(99)80026-4.
Arshad T, Khan KM, Rasool N, Salar U, Hussain S, Asghar H, et al. 5-Bromo-2-aryl benzimidazole derivatives as non-cytotoxic potential dual inhibitors of α-glucosidase and urease enzymes. Bioorg Chem. 2017;72:21–31. https://doi.org/10.1016/j.bioorg.2017.03.007.
Hanif M, Shoaib K, Saleem M, Hasan Rama N, Zaib S, Iqbal J. Synthesis, urease inhibition, antioxidant, antibacterial, and molecular docking studies of 1, 3, 4-oxadiazole derivatives. ISRN Pharmacol. 2012;2012. https://doi.org/10.5402/2012/928901.
Horta LP, Mota YC, Barbosa GM, Braga TC, Marriel IE, Fátima ÂD, et al. Urease inhibitors of agricultural interest inspired by structures of plant phenolic aldehydes. J Braz Chem Soc. 2016;27:1512–9. https://doi.org/10.21577/0103-5053.20160208.
Hakimi AM, Lashgari N, Mahernia S, Ziarani GM, Amanlou M. Facile one-pot four-component synthesis of 3, 4-dihydro-2-pyridone derivatives: novel urease inhibitor scaffold. Res Pharm Sci. 2017;12:353 https://doi.org/10.4103/1735-5362.213980.
Svane S, Sigurdarson JJ, Finkenwirth F, Eitinger T, Karring H. Inhibition of urease activity by different compounds provides insight into the modulation and association of bacterial nickel import and ureolysis. Sci Rep. 2020;10:1–4. https://doi.org/10.1038/s41598-020-65107-9.
Rego YF, Queiroz MP, Brito TO, Carvalho PG, de Queiroz VT, de Fátima Â, et al. A review on the development of urease inhibitors as antimicrobial agents against pathogenic bacteria. J Adv Res. 2018;13:69–100. https://doi.org/10.1016/j.jare.2018.05.003.
Kafarski P, Talma M. Recent advances in design of new urease inhibitors: A review. J Adv Res. 2018;13:101–12. https://doi.org/10.1016/j.jare.2018.01.007.
Ansari FL, Wadood A, Ullah A, Iftikhar F, Ul-Haq Z. In silico studies of urease inhibitors to explore ligand-enzyme interactions. J Enzym Inhib Med Chem. 2009;24:151–6. https://doi.org/10.1080/14756360801945598.
Schneider A, Garlick JA, Egles C. Self-assembling peptide nanofiber scaffolds accelerate wound healing. PloS one. 2008;3:1410 https://doi.org/10.1371/journal.pone.0001410.
Meng H, Chen L, Ye Z, Wang S, Zhao X. The effect of a self‐assembling peptide nanofiber scaffold (peptide) when used as a wound dressing for the treatment of deep second degree burns in rats. J Biomed Mater Res Part B. 2009;89:379–91. https://doi.org/10.1002/jbm.b.31226.
Koutsopoulos S. Self‐assembling peptide nanofiber hydrogels in tissue engineering and regenerative medicine: Progress, design guidelines, and applications. J Biomed Mater Res Part A. 2016;104:1002–16. https://doi.org/10.1002/jbm.a.35638.
Tang C, Shao X, Sun B, Huang W, Zhao X. The effect of self-assembling peptide RADA16-I on the growth of human leukemia cells in vitro and in nude mice. Int J Mol Sci. 2009;10:2136–45. https://doi.org/10.3390/ijms10052136.
Mi K, Wang G, Liu Z, Feng Z, Huang B, Zhao X. Influence of a self‐assembling peptide, RADA16, compared with collagen I and matrigel on the malignant phenotype of human breast‐cancer cells in 3D cultures and in vivo. Macromol Biosci. 2009;9:437–43. https://doi.org/10.1002/mabi.200800262.
Fung SY, Yang H, Chen P. Formation of colloidal suspension of hydrophobic compounds with an amphiphilic self-assembling peptide. Colloids Surf B. 2007;55:200–11. https://doi.org/10.1016/j.colsurfb.2006.12.002.
Wang M, Adikane HV, Duhamel J, Chen P. Protection of oligodeoxynucleotides against nuclease degradation through association with self-assembling peptides. Biomaterials. 2008;29:1099–108. https://doi.org/10.1016/j.biomaterials.2007.10.049.
Liu J, Zhang L, Yang Z, Zhao X. Controlled release of paclitaxel from a self-assembling peptide hydrogel formed in situ and antitumor study in vitro. Biomaterials 2011;6:2143 https://doi.org/10.2147/IJN.S24038.
Zhang H, Xin X, Sun J, Zhao L, Shen J, Song Z, et al. Self-assembled chiral helical nanofibers by amphiphilic dipeptide derived from d-or l-threonine and application as a template for the synthesis of Au and Ag nanoparticles. J Colloid Interface Sci. 2016;484:97–106. https://doi.org/10.1016/j.jcis.2016.08.052.
Merrifield RB. Solid phase peptide synthesis. I. The synthesis of a tetrapeptide. J Am Chem Soc. 1963;85:2149–54. https://doi.org/10.1021/ja00897a025.
Balasubramanian A, Ponnuraj K. Crystal structure of the first plant urease from jack bean: 83 years of journey from its first crystal to molecular structure. J Mol Biol. 2010;400:274–83. https://doi.org/10.1016/j.jmb.2010.05.009.
Taha M, Ismail NH, Khan A, Shah SA, Anwar A, Halim SA, et al. Synthesis of novel derivatives of oxindole, their urease inhibition and molecular docking studies. Bioorg Med Chem Lett. 2015;25:3285–9. https://doi.org/10.1016/j.bmcl.2015.05.069.
Funding
Thanks to Higher Education Commission (HEC), Pakistan, for providing SRGP project (NO: 21-1505/SRGP/R&D/HEC/2017) and 8169 /Sindh/NRPU/R&D/HEC/2017 from the Higher Education Commission, Pakistan.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Shah, Z.A., Hussain, S., Khan, S. et al. Inhibition of jack bean urease by amphiphilic peptides. Med Chem Res 30, 1569–1576 (2021). https://doi.org/10.1007/s00044-021-02757-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00044-021-02757-y