- 1Department of Clinical Laboratory, Xiangya Hospital, Central South University, Changsha, Hunan, China
- 2National Clinical Research Center for Geriatric Disorders, Xiangya Hospital, Changsha, Hunan, China
Purpose: The type VI system (T6SS) has the potential to be a new virulence factor for hypervirulent Klebsiella pneumoniae (hvKp) strains. This study aimed to characterize the molecular and clinical features of T6SS-positive and T6SS-negative K. pneumoniae isolates that cause abscesses.
Patients and methods: A total of 169 non-duplicate K. pneumoniae strains were isolated from patients with abscesses in a tertiary hospital in China from January 2018 to June 2022, and clinical data were collected. For all isolates, capsular serotypes, T6SS genes, virulence, and drug resistance genes, antimicrobial susceptibility testing, and biofilm formation assays were assessed. Multilocus sequence typing was used to analyze the genotypes of hvKp. T6SS-positive hvKp, T6SS-negative hvKp, T6SS-positive cKP, and T6SS-negative cKP (n = 4 strains for each group) were chosen for the in vivo Galleria mellonella infection model and in vitro competition experiments to further explore the microbiological characteristics of T6SS-positive K. pneumoniae isolates.
Results: The positive detection rate for T6SS was 36.1%. The rates of hvKp, seven virulence genes, K1 capsular serotype, and ST23 in T6SS-positive strains were all higher than those in T6SS-negative strains (p < 0.05). Multivariate logistic regression analysis indicated that the carriage of aerobactin (OR 0.01) and wcaG (OR 33.53) were independent risk factors for T6SS-positive strains (p < 0.05). The T6SS-positive strains had a stronger biofilm-forming ability than T6SS-negative strains (p < 0.05). The T6SS-positive and T6SS-negative strains showed no significant differences in competitive ability (p = 0.06). In the in vivo G. mellonella infection model, the T6SS(+)/hvKP group had the worst prognosis. Except for cefazolin and tegacyclin, T6SS-positive isolates displayed a lower rate of antimicrobial resistance to other drugs (p < 0.05). The T6SS-positive isolates were more likely to be acquired from community infections (p < 0.05).
Conclusion: Klebsiella pneumoniae isolates causing abscesses have a high prevalence of T6SS genes. T6SS-positive K. pneumoniae isolates are associated with virulence, and the T6SS genes may be involved in the hvKp virulence mechanism.
1. Introduction
Klebsiella pneumoniae is a threat to public health and can cause a range of severe infections, such as pneumonia, septicemia, meningitis, urinary tract infection, and bacterial abscesses (Dong et al., 2019; Lai and Yu, 2021). Based on its virulence features, K. pneumoniae is commonly classified into two groups: classic K. pneumoniae (cKp) and hypervirulent K. pneumoniae (hvKp) (Liu and Guo, 2018). CKp exhibits low virulence and with high resistance to antibiotics, which primarily causes pneumoniae, urinary tract infection and bacteremia in patients with severe underlying diseases or immunocompromised (Zhao et al., 2021; Kamau et al., 2022). Currently, hvKp has drawn extensive attention from researchers and clinicians, as it induces invasive abscess infections, especially liver abscesses, and has high pathogenicity resulting in high mortality (Zhu et al., 2021; Lin et al., 2022). Thus, knowledge of the underlying pathogenesis of hvKp is important for targeted disease prevention and control.
Protein secretion apparatuses play critical roles in bacterial survival and adaptation to complex environments and are used by bacteria to transport proteins through their cell membranes into external host cells (Morra et al., 2018). Currently, nine secretion systems are known: T1SS, T2SS, T3SS, T4SS, T5SS, T6SS, T7SS, T8SS, and T9SS (Hui et al., 2021). The type VI system (T6SS) is widespread in gram-negative bacteria (Yu et al., 2021), and in K. pneumoniae, it promotes bacterial competition, cell invasion, type-1 fimbriae expression, and in vivo colonization (Hsieh et al., 2019).
Strong iron uptake is one of the key features of hvKp (Choby et al., 2020; Nicolo et al., 2022). Genomic analysis identified that several genes coding for iron uptake are encoded around the T6SS gene; thus, T6SS could be involved in iron uptake in hvKp and may be a new virulence factor in hvKp strains (Barbosa and Lery, 2019; Zhu et al., 2021; Altayb et al., 2022). Two studies reported the frequency of the T6SS gene in K. pneumoniae isolated from bloodstream infections (Zhou et al., 2020; Zhang et al., 2022). However, the incidence rate of the T6SS gene in K. pneumoniae causing abscesses and in vivo studies of T6SS virulence has not been reported.
Here, we chose clinical K. pneumoniae-induced abscesses which usually have a high incidence of hvKp to obtain further insight into the T6SS relationships with hvKp. This study assessed the distribution of T6SS genes and characterized the molecular and clinical features of T6SS-positive and T6SS-negative isolates to further investigate the connection between T6SS and hypervirulence and to better understand the virulence mechanisms of hvKp.
2. Materials and methods
2.1. Isolates and clinical data collection
We isolated 169 non-duplicate K. pneumoniae strains from patients with abscesses in a tertiary hospital in China from January 2018 to June 2022 and stored them at −80°C. K. pneumoniae isolates were identified using matrix-assisted laser desorption/ionization-time of flight mass spectrometry (Bruker Daltonics). Abscess infections in which the detection of bacteria occurred in ≤48 h after admission were defined as community-acquired, while those that took >48 h after admission and infections associated with medical devices were classified as hospital-acquired (Anderson et al., 2017).
In addition, the following clinical information was collected from the patients’ electronic medical records: age, sex, acquisition of infection, personal history, underlying disease, antibiotic use, invasive operations, and clinical outcomes.
2.2. Hyperviscosity phenotype
The hyperviscosity phenotype was detected using the string test. The strains were cultured on blood agar plates, and then a single colony was gently picked with an inoculation loop. If a viscous string with a length > 5 mm could be pulled out, the strain was considered to have a hyperviscous phenotype. The tests were repeated ≥3 times.
2.3. Detection of T6SS, capsular serotyping, virulence, and drug resistance genes
The presence of T6SS, capsular serotyping, virulence, and drug resistance genes was detected by polymerase chain reaction (PCR), as previously described (Zhou et al., 2020). Genomic DNA of K. pneumoniae was extracted using the boiling method. The PCR products were electrophoresed on 1.0% agarose gel at 100 V for 40 min and visualized using a Syngene gel imager to determine the presence of these genes. In this study, strains positive for p-rmpA, rmpA2, and aerobactin were defined as hvKP, and those all positive for IcmF, vgrG, and hcp were designated as T6SS-positive strains. The primers used in this study are listed in the Supplementary material.
2.4. Whole-genome sequencing and multilocus sequence typing (MLST)
Whole-genome sequencing was used to identify virulence genes and sequence types (STs) in 70 hvKp isolates. Genomic DNA was extracted using the Ezup Column Bacteria Genomic DNA Purification Kit (Sangon Biotech, Shanghai, China). After sample qualification and library preparation, the sequencing library was sequenced on a DNBSEQ-T7 with the PE150 model at Bioyi Biotechnology Co., Ltd. (Wuhan, China). The data were assembled using Unicycler (v0.4.8) (Wick et al., 2017) and the whole genome was obtained. STs were identified using the MLST database.1 The 70 hvKp isolates were aligned to the reference genome, SGH10 K. pneumoniae (CP025080), using Snippy software (v4.6.0), and the core gene single nucleotide polymorphism sites were extracted. After removing the recombinant regions using the Gubbins software, phylogenetic trees using the maximum likelihood method were constructed using the FastTree software and uploaded to the iTOL website2 for annotation. The sequence data were deposited in the National Center for Biotechnology Information database with the accession numbers PRJNA940638. Using as a reference sequence.
2.5. Biofilm formation assay
The 0.5 McFarland bacterial suspensions were diluted (1:100), and 200 μL of the suspension was pipetted into a 96-well plate (three parallel wells were set up for each sample), and the microtiter plate was incubated for 24 h at 37°C. After pouring out the bacterial solution, each well was gently rinsed (3×) with 200 μL phosphate-buffered saline and air-dried. Subsequently, each well was stained with 200 μL of 1% crystalline violet for 15 min, the excess dye was washed in the wells and left to dry in the room, and 200 μL of 95% ethanol was added to each well to dissolve the crystal violet. After 10 min, the absorbance values were measured at 570 nm three times to obtain an average value. The mean OD570 value of the blank control plus 3× standard deviation was used as the ODc. Biofilm formation was defined as negative, weak, moderate, and strong if OD ≤ ODc, ODc < OD ≤ 2ODc, 2ODc < OD ≤ 4ODc, and 4ODc < OD, respectively.
2.6. In vitro competition assay
In vitro competition assays were performed as described previously, with slight modification (Zhang et al., 2022). K. pneumoniae isolates and Escherichia coli strain 25,922 were cultured overnight at 37°C in liquid Luria-Bertani (LB) medium. After standardization to an OD600 of 0.5, each culture was diluted (1:100) in 10 mL LB liquid medium. Suspensions of K. pneumoniae and E. coli were mixed at a ratio of 2:1 and incubated at 180 rpm and 37°C for 20 h. Serial 10-fold dilutions were applied to LB plates with or without ampicillin (50 μg/mL) and the cell counts of each strain were determined. The relative competitive index (CI) was calculated as the ratio of ampicillin-resistant K. pneumoniae to ampicillin-susceptible E. coli.
2.7. In vivo Galleria mellonella infection model
An in vivo virulence assay was performed using the G. mellonella infection model. Ten G. mellonella larvae weighing 250–350 mg were used for each strain (purchased from Tianjin Huiyude Biotechnology). Bacterial suspensions (10 μL; 1 × 106 colony forming unit [CFU]) were inoculated into the left hind foot, and the number of deaths was recorded every 12 h and observed continuously for 72 h. All experiments were performed three times.
2.8. Antimicrobial susceptibility testing
Antimicrobial susceptibility testing was performed using the VITEK®2 Compact system (bioMerieux, Marcy l’Etoile, France) according to the manufacturer’s instructions. A panel of 20 antimicrobial agents was identified on the gram-negative Bacillus drug sensitivity identification card. K. pneumoniae ATCC700603 and E. coli ATCC25922 were used as the quality control strains. Bacterial resistance was determined based on minimal inhibitory concentrations (MIC) according to the Clinical and Laboratory Standards Institute 2022 standard (Lewis, 2022).
2.9. Statistical analysis
Categorical variables were analyzed using the chi-squared test or Fisher’s exact probability method. Student’s t-test or Mann–Whitney U-test was used to analyze numerical variables. p-value <0.05 was considered statistically significant. Virulence, resistance, and capsular serotyping genes, and clinical characteristics were analyzed by one-way logistic regression and multi-way logistic regression to determine risk factors for T6SS-positive K. pneumoniae infection of abscess origin. All data analyses were performed using the R4.2.1.
2.10. Ethics statement
Approval to collect patient medical records (which were anonymized) and K. pneumoniae strains was granted by the Ethics Committee of Xiangya Hospital, Central South University. Informed consent was not obtained due to the retrospective nature of the study. This study was also approved by the Central South University Ethics Committee (ID 202212281; Changsha, Hunan Province, People’s Republic of China).
3. Results
3.1. Clinical characteristics
The average age of all patients was 52.41 (46.00–63.00) years, and 71.01% (120/169) were men (Table 1); there were no obvious differences in age or sex between the two groups. The most common abscess site was the liver (33.14%), followed by the skin (12.43%). The most common disease was diabetes (33.73%), followed by liver and gallbladder diseases (32.54%), and lung diseases (31.95%). Most patients had antibiotic exposure before diagnosis (83.43%), and a few patients had multiple bacterial infections (37.28%). Among the T6SS-positive patients, the prevalence of hyperlipidemia was high (p < 0.05). Moreover, there were more community infections in the T6SS-positive group than in the T6SS-negative group (p < 0.05), and there were no significant differences in age, sex and other clinical characteristics (p > 0.05).
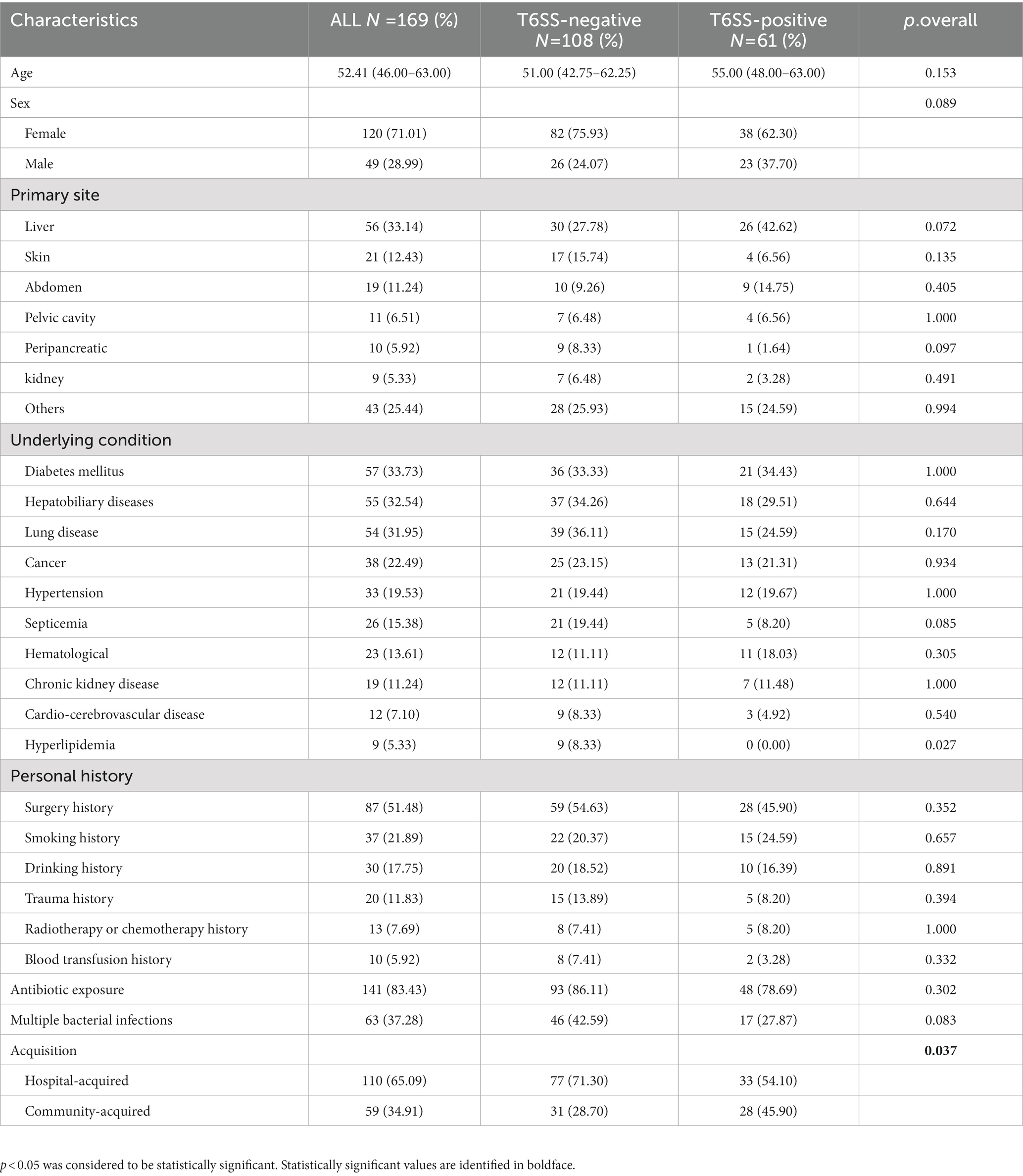
Table 1. Clinical characteristics of type VI secretion system (T6SS)-positive and T6SS-negative Klebsiella pneumoniae isolates causing abscesses.
3.2. Distribution of T6SS, virulence- and drug-resistance genes, capsular serotypes, and hyperviscosity phenotype
Table 2 compares the distribution of hyperviscosity phenotype, virulence and drug resistance genes, and capsular types between T6SS-positive and T6SS-negative K. pneumoniae isolates. We tested the prevalence of 11 virulence genes: rmpA, rmpA2, aerobactin, iroB, peg344, mrkD, wcaG, ybtS, alls, Kfu, entB, and iucA. The positive rates of rmpA, rmpA2, aerobactin, iroB, wcaG, all, and Kfu were significantly higher in the T6SS-positive K. pneumoniae strains than in the T6SS-negative isolates (p < 0.05). In contrast, the positive rates of K. pneumoniae carbapenemase (KPC) and KPC-2 drug resistance genes in the T6SS-negative K. pneumoniae isolates were significantly higher than those in the T6SS-positive K. pneumoniae isolates (p < 0.001). As determined by p-rmpA, iroB, and iucA, 81 strains (47.65%) were identified as hvKp. Among T6SS-positive strains, the proportion of hvKp was significantly high (p < 0.001). Among the 169 K. pneumoniae strains, capsular types K1, K2, and K64 accounted for 25.88% (44/170), 21.18% (36/170), and 11.76% (20/170), respectively. T6SS-positive K.pneumoniae strains were observed to have a significant high prevalence for K1 serotype and T6SS-negative K.pneumoniae isolates have a significant high prevalence for K2 serotype (p < 0.001).
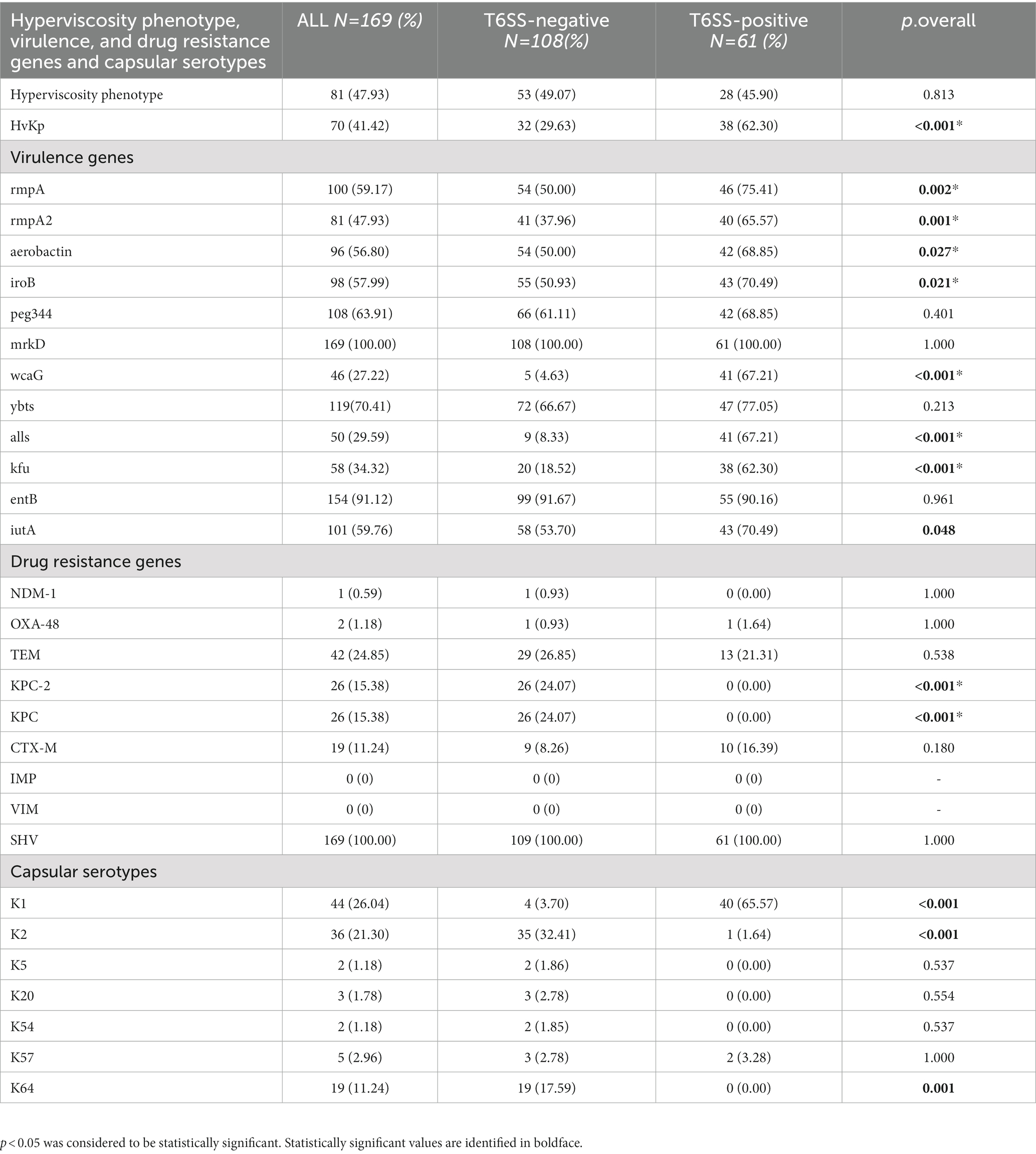
Table 2. Hyperviscosity phenotype, virulence, and drug resistance genes and capsular serotypes distribution of T6SS-positive and T6SS-negative K. pneumoniae isolates causing abscesses.
3.3. MLST and phylogenic analysis
As shown in Table 3 and the core genome phylogenetic tree (Figure 1), similar to other studies, the most common ST in hvKp was ST23 (51.43%), and the most common serotype in hvKp was K1 (54.29%). Among the 36 ST23 strains, 16 had K1 capsules. From 2018 to 2022, there were 10,13,19, 17, and 11 hvKp strains. In T6SS-positive hvKp, the positivity rate of ST23 was 92.11%, which was higher than that of T6SS-negative hvKp (3.12%). In addition, the positive rates of ST65 and ST86 in T6SS-positive hvKp were far lower than that in T6SS-negative hvKp.
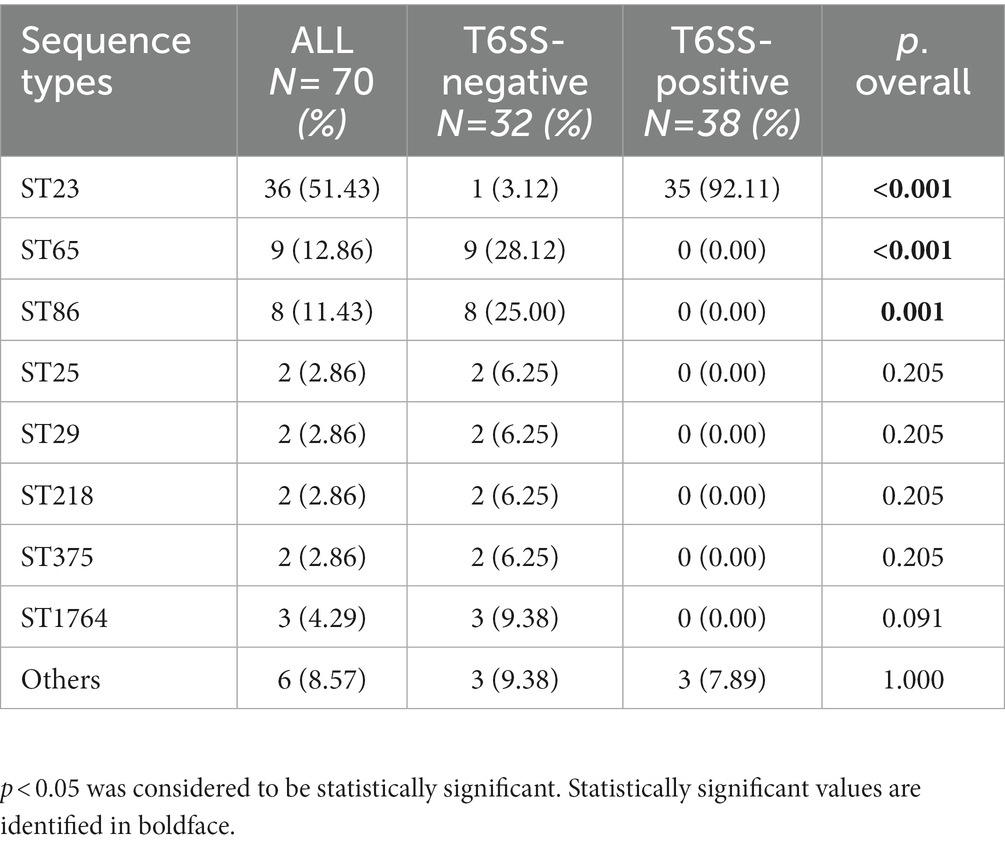
Table 3. Multilocus sequence typing of T6SS-positive and T6SS-negative K. pneumoniae isolates causing abscesses.
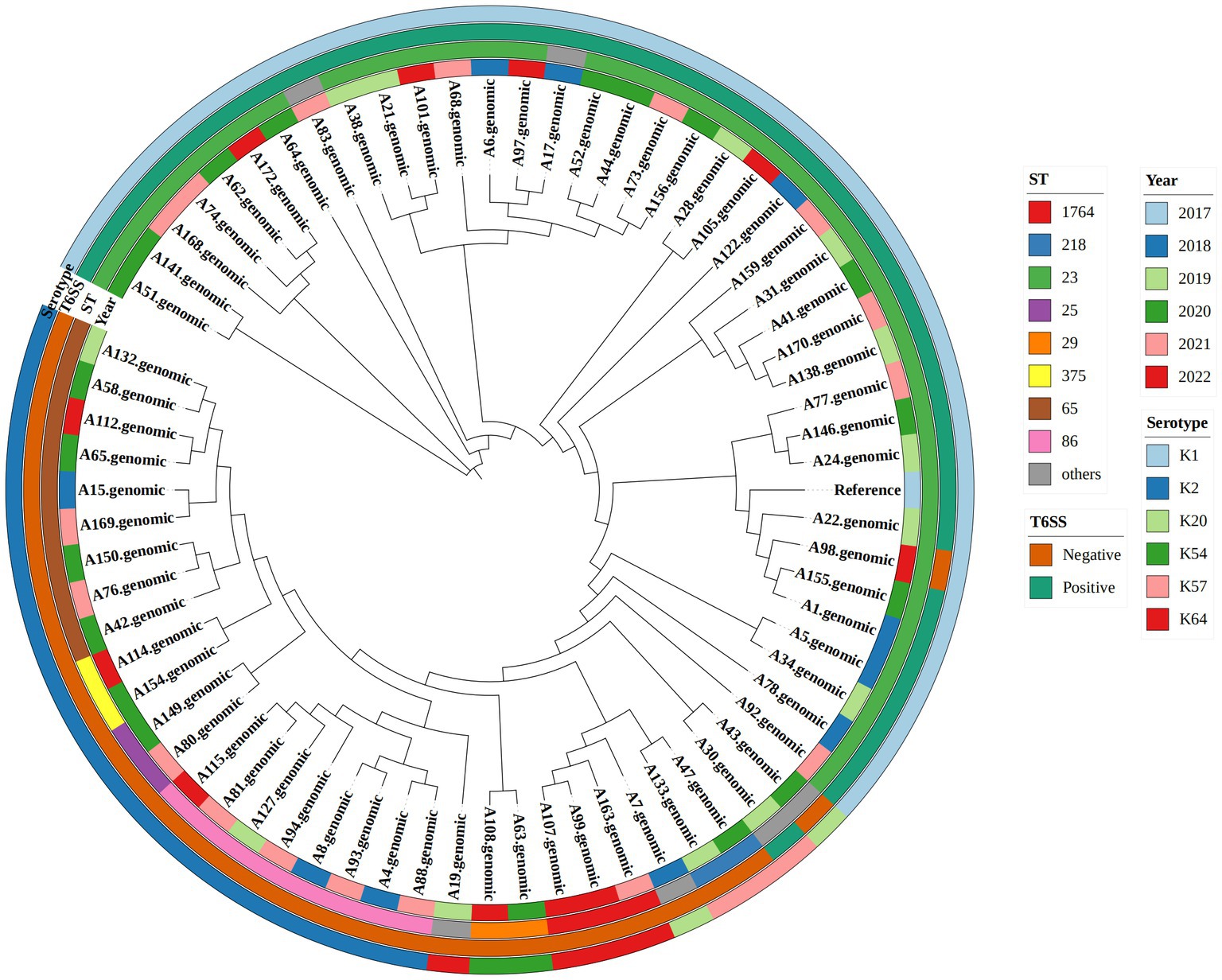
Figure 1. Phylogenetic tree of 70 hypervirulent Klebsiella pneumoniae isolates. From outside to center, rings 1, 2, 3, and 4 show the serotype, T6SS gene, year, and sequence type, respectively.
3.4. Antimicrobial susceptibility
Among the 169 K. pneumoniae isolates, in addition to natural resistance to ampicillin, K. pneumoniae had the highest resistance to cefazolin (36.09%), and the resistance rate to tigecycline was as low as 1.78% (Table 4). The drug resistance of all T6SS-positive K. pneumoniae was lower than that of the T6SS-negative strains. Except for cefazolin and tigecyline, the drug sensitivity results for the other drugs were significantly different (p < 0.05). In addition, K. pneumoniae with K1 serotype showed lower drug resistance to 19 drugs except for ampicillin (p < 0.05) and K. pneumoniae with K2 and K57 serotype showed lower drug resistance to 10 drugs and 8 drug, respectively (p < 0.05). In contrast, K. pneumoniae with K64 serotype showed higher drug resistance to 18 drugs except for ampicillin and tigecycline (p < 0.05). The detailed results were given in the Supplementary material.
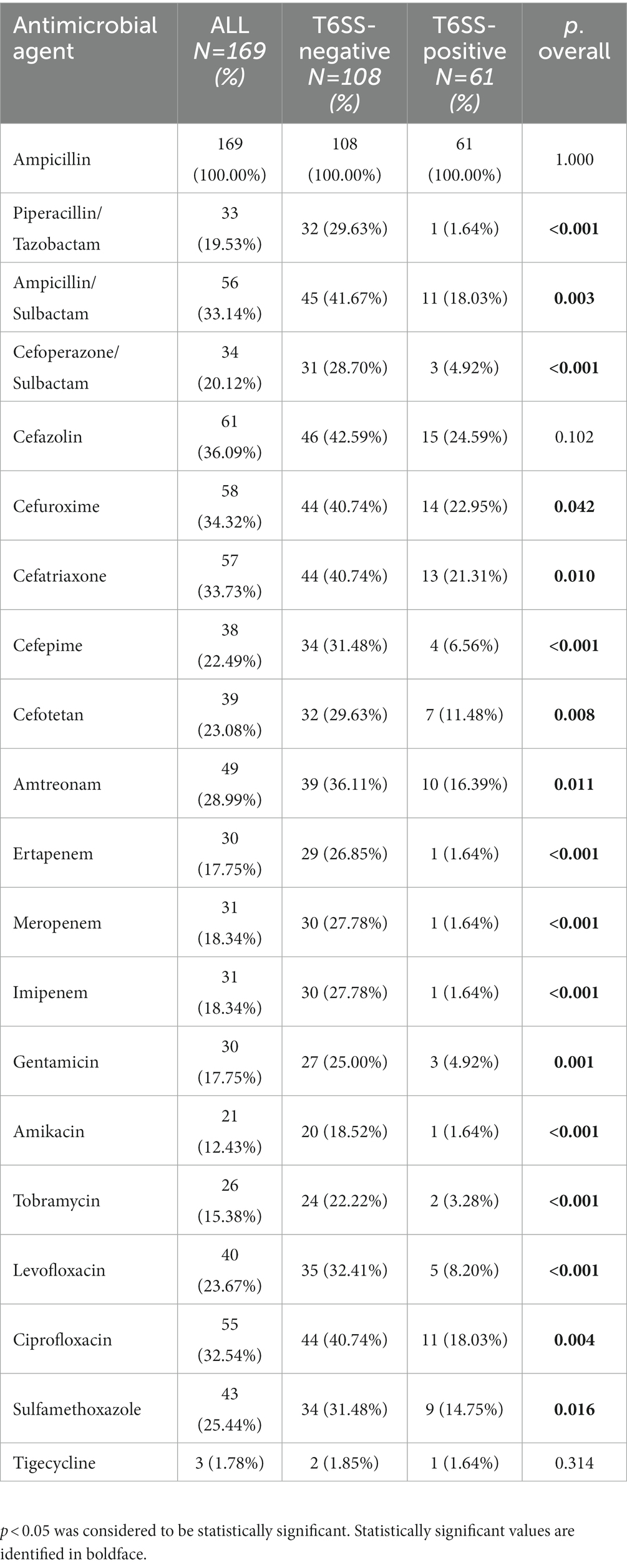
Table 4. Antimicrobial resistance of T6SS-positive and T6SS-negative K. pneumoniae isolates causing abscesses.
3.5. Risk factor analysis
After single-factor logistic regression of virulence genes, drug resistance genes and clinical characteristics were included, and the results showed that the following nine genes were significantly related to T6SS-positive strains infection: rmpA, rmpA2, aerobactin, iroB, wcaG, all, Kfu, K1 and K2. In addition, hvKp and community-acquired infections were significantly associated with T6SS-positive strain infection. The above nine variables were included in multi-factor logistic regression (Figure 2); aerobactin(OR 0.01) and WcaG (OR = 33.53)are independent risk factor for T6SS-positive strains infection (p < 0.05).
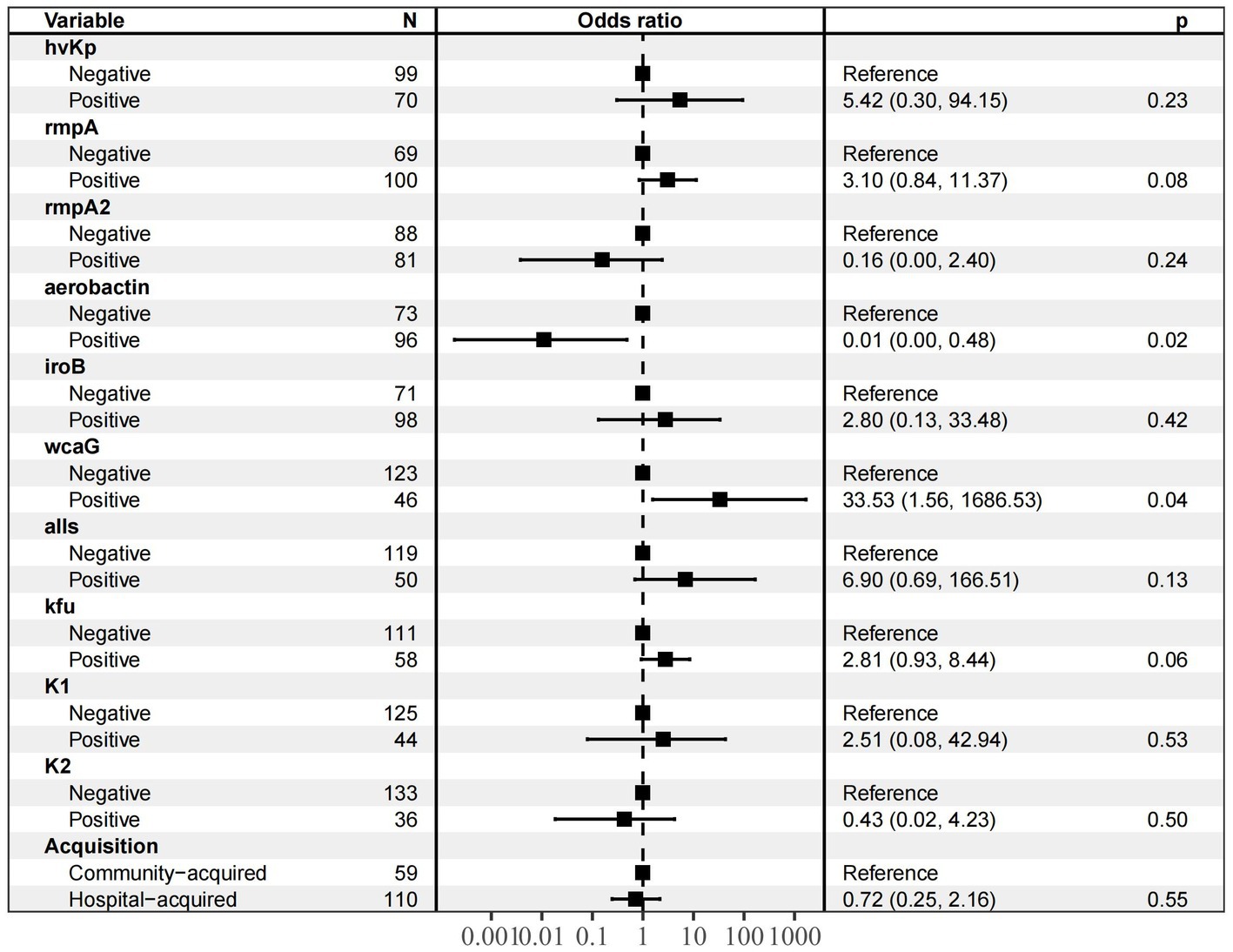
Figure 2. Multivariate analysis of prognostic factors for patients with T6SS − positive and T6SS-negative K.pneumoniae abscesses infections. The nine variables (hvKp, rmpA, rmpA2, aerobactin, iroB, wcaG, alls, Kfu, K1, K2 and acquisiyion) were included in multi-factor logistic regression, and aerobactin (OR 0.01) and WcaG (OR = 33.53)are independent risk factor for T6SS-positive strains infection (p < 0.05).
3.6. Biofilm formation
Twenty-eight (16.57%), 74 (43.79%), and 67 (39.64%) strains had strong, medium, and weak biofilm-forming ability, respectively. As shown in Table 5; Figure 3, the proportion of isolates with strong biofilm-forming ability among T6SS-positive strains was higher than that of T6SS-negative strains (p < 0.05). Also, in different serotypes of K. pneumoniae strains only K64’s biofilm formation was statistically significant, and it had lower biofilm formation (p < 0.05). The detailed results were given in the Supplementary material.
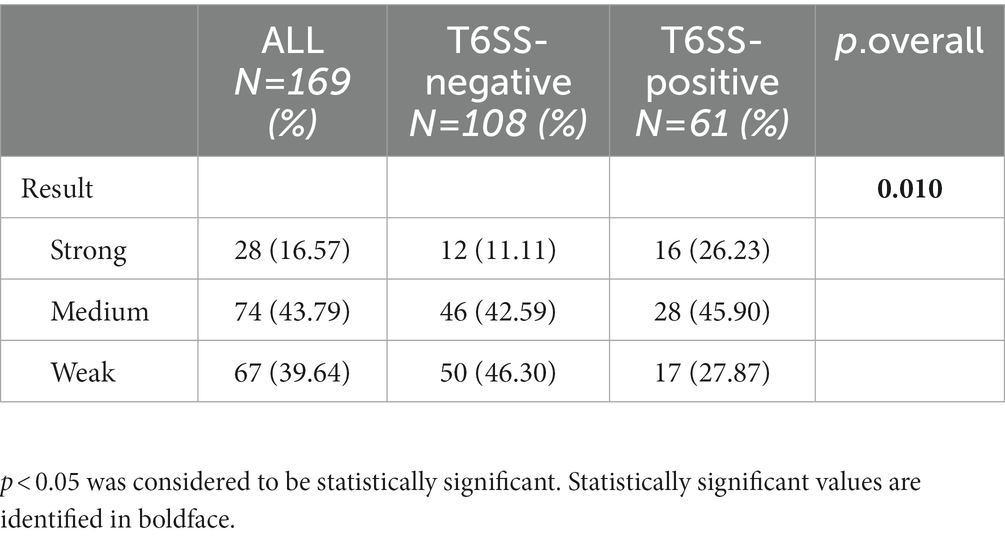
Table 5. Biofilm formation ability of T6SS-positive and T6SS-negative K. pneumoniae isolates causing abscesses.
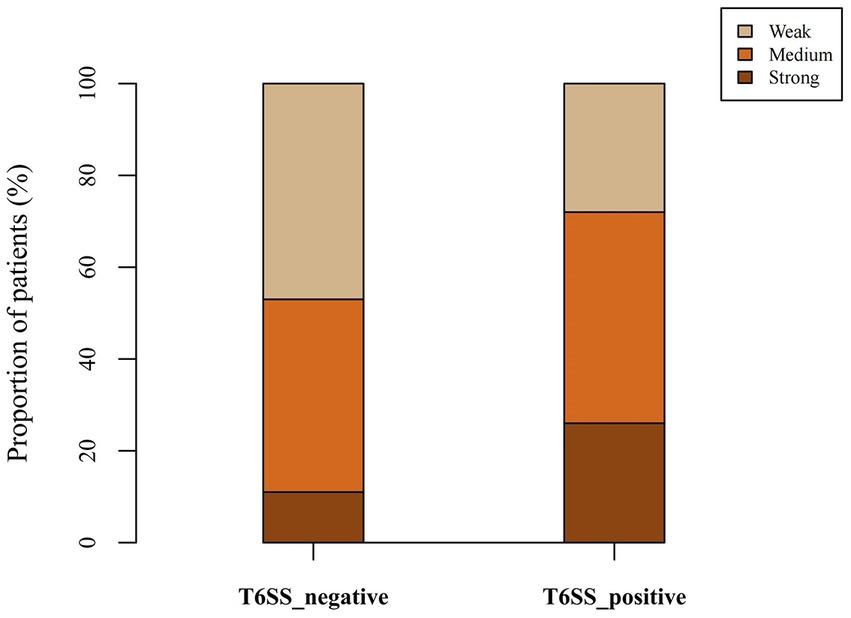
Figure 3. Proportion of biofilm-forming ability of T6SS-positive and T6SS-negative K. pneumoniae isolates causing abscesses. The proportion of isolates with strong biofilm-forming ability among T6SS-positive strains was higher than that of T6SS-negative strains.
3.7. In vitro competition experiments
This study calculated the CI values of 16 K. pneumoniae strains which were divided into four groups of four T6SS-positive and hvKp (T6SS[+]/hvKP), T6SS-positive and cKP (T6SS[+]/cKP), T6SS-negative and hvKp (T6SS[−]/hvKP), and T6SS-negative and cKP (T6SS[−]/cKP) strains (Figure 4). The average CI values of the T6SS(+)/hvKP, T6SS(+)/cKP, T6SS(−)/hvKP, and T6SS(−)/cKP were 1.36, 1.16, 1.38, and 2.34, respectively. The results showed that during co-culture with E. coli, there was no difference between the competitiveness of T6SS-positive and T6SS-negative strains (p = 0.06), and there was no difference between the competitiveness of cKP and hvKP strains (p = 0.25).
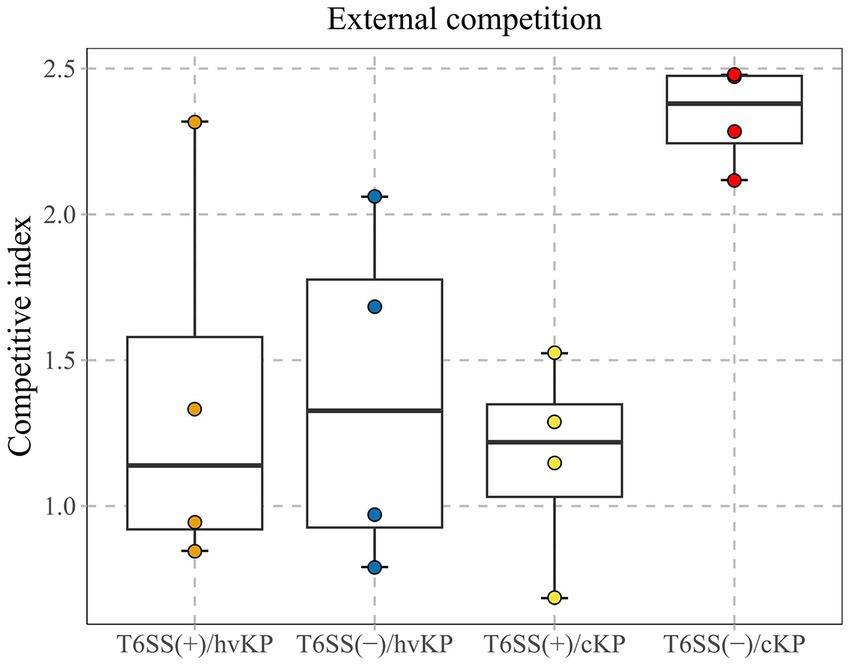
Figure 4. ln vitro competitive index (CI) values of T6SS K. pneumoniae isolates causing abscesses. Each circle represents one of the Cl values obtained from the number of ampicillin-resistant CFUs (K. pneumoniae) divided by the number of ampicillin-susceptible CFUs (E. coli). There was no difference between the competitiveness of T6SS-positive and T6SS-negative strains (p = 0.06), and there was no difference between the competitiveness of cKP and hvKP strains (p = 0.25).
3.8. In vivo survival analysis of Galleria mellonella infection model
We selected 16 K. pneumoniae strains for further analysis using an in vivo G. mellonella infection model and assessed the different groups as described above (section 3.7). The T6SS(+)/hvKP group had the worst prognosis, and the survival rate of this group was significantly different from the other three groups (p < 0.05; Figure 5).
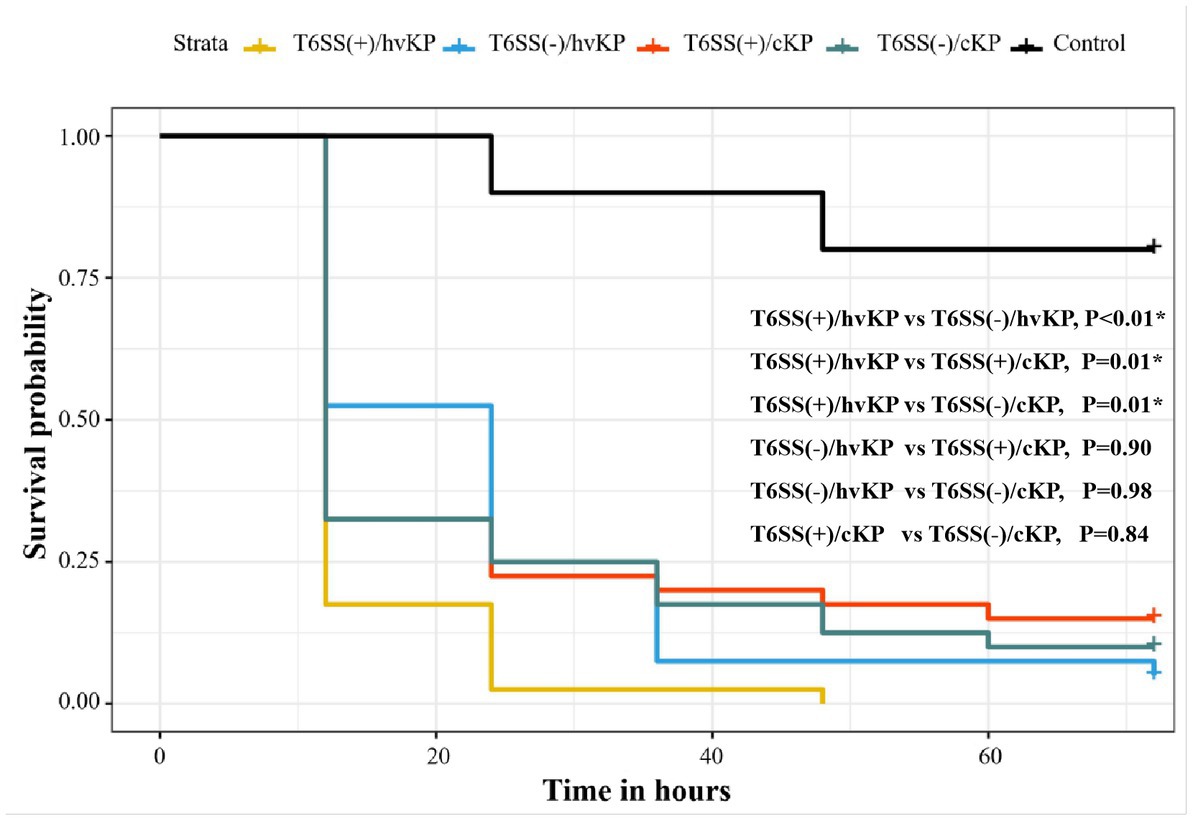
Figure 5. Evaluation of the virulence of K. pneumoniae isolates causing abscesses. Using a G. mellonella infection model, we investigated the virulence of 1 × 106 CFU of each group of T6SS(+)/hvKP, T6SS(−)/hvKP, T6SS(+)/cKP, and T6SS(−)/cKP strains. Normal saline was used as control. The T6SS(+)/hvKP group had the worst prognosis, and the survival rate of this group was significantly different from the other three groups (p < 0.05).
4. Discussion
This study analyzed the molecular epidemiological characteristics of T6SS system genes in K. pneumoniae causing abscesses and the clinical features of 169 patients from January 2018 to June 2022. Studies that specifically investigate invasive abscess infections of K. pneumoniae isolates are rare.
Positive results for hcp, icmF, and vgrG genes confirmed T6SS positivity (Zhou et al., 2020; Liao et al., 2022; Zhang et al., 2022), and 36.1% (61/169) of our strains were T6SS-positive; this was much higher than the rate in blood stream infection (BSI) (20.1%) (Zhou et al., 2020). We identified hvKp in 41.42% (70/169) of patients and hypermucoviscosity phenotype in 47.93% (81/169). Moreover, the results indicated that there were more hvKp strains among T6SS-positive strains than among T6SS-negative strains (p < 0.001), which was similar to the findings of a study by Yin et al. (Zhang et al., 2022). The most prevalent hvKp isolates were ST23 (36/70), and ST23 in T6SS-positive isolates was significantly higher than in the T6SS-negative isolates. ST23 is closely related to high virulence, and K. pneumoniae ST23 is considered an important pathogen that causes hospital-acquired infections in critical patients (Lee et al., 2017; Hernandez et al., 2021; Du et al., 2022; Fan et al., 2022). These results suggest that the T6SS-positive K. pneumoniae strains are highly virulent.
However, there was no significant difference in the hypermucoviscosity phenotype between the T6SS-positive and T6SS-negative isolates. Previous studies have traditionally used the hypermucoviscosity phenotype to identify hvKp (Zhang et al., 2016). But increasing evidence has shown that hvKp may not be fully judged by hypermucoviscosity, and it is only one of the phenotypes that hvKp possesses (Liu and Guo, 2019; Su et al., 2020; Walker et al., 2020). Not all hvKp strains possessed the hypermucoviscosity phenotype in our study, and vice versa.
We investigated 11 virulence-associated and nine drug resistance-associated genes and found that the positive rates of rmpA, rmpA2, aerobactin, iroB, wcaG, alls, and Kfu were relatively higher for T6SS-positive isolates than for T6SS-negative isolates (p < 0.001). These seven virulence factors are associated with high pathogenicity: rmpA and rmpA2 are involved in the hypermucoviscosity phenotype and capsular polysaccharide synthesis; aerobactin and iroB are siderophores, and aerobactin plays a critical role in hvKp virulence; and wcaG regulates fucose biosynthesis and enhances the ability of K. pneumoniae to resist neutrophil phagocytosis (Zheng et al., 2018; Russo and Gulick, 2019; Liu et al., 2022; Xu et al., 2022). These results further indicate that the T6SS-positive isolates were hypervirulent; however, the specific virulence mechanism involved in this process requires further study.
The T6SS-negative isolates displayed a higher rate of drug-resistant genes, KPC-2 and KPC, than T6SS-positive isolates (p < 0.001). KPC is a carbapenemase gene, and the presence of it could contribute the resistance to carbapenems (ertapenem, imipenem and meropenem) in K. pneumoniae (Effah et al., 2020; Cienfuegos-Gallet et al., 2022). Compared to the T6SS-positive isolates that all without KPC or KPC-2 gene, 24.07%(26/108) of T6SS-negative isolates had KPC and KPC-2 gene, and these T6SS-negative isolates all showed resistance to ertapenem, imipenem and meropenem. Antimicrobial susceptibility analysis revealed that the majority of T6SS-positive strains were sensitive to all antimicrobials except for ampicillin, similar to hvKp strains that are usually sensitive to antibiotics (Gu et al., 2018). Antibiotic resistance is often associated with decreased virulence (Mei et al., 2017). Hence, to a certain degree, this result shows that the T6SS genes may be linked to virulence.
K1 and K2 were strongly associated with high virulence in the previous study, and our results also found that K1 and K2 showed lower drug resistance (Lee et al., 2017; Sohrabi et al., 2022). In our study, T6SS-positive strains had a higher detection rate of the K1 capsular serotype than T6SS-negative strains. However, unlike other studies, the detection rates of K2 and K64 capsular serotypes in T6SS-negative strains were significantly higher than those of T6SS-positive strains (p < 0.05). Although little has been studied about the strength and weakness of the virulence of K1 and K2 serotypes, some studies have shown that K1 is more predominant in hvKp than K2 (Liu et al., 2014; Liu and Guo, 2018; Sohrabi et al., 2022). According to our result, it may also suggest that K1 were more likely associated with hvKp compared with K2. More studies are required to confirm this. In our study K64 showed higher drug resistance and it was reported that K64 was commonly associated with carbapenem resistance (Wang et al., 2022). The main mechanism of carbapenem resistance include enzyme production, efflux pumps and porin mutations (Suay-Garcia and Perez-Gracia, 2019). Our result found that K64 with higher drug resistance showed lower biofilm formation which suggested K64’s drug resistance mechanism probably not related to biofilm formation.
Patients infected with T6SS-positive isolates presented with clinical features similar to those of hvKp. Clinical characteristic analysis showed that community-acquired infections were more common in patients infected with T6SS-positive isolates than in those infected with T6SS-negative isolates. HvKp has often caused serious invasive community-acquired infections in previous studies (Wyres et al., 2020; Lam et al., 2021).
Our in vivo pathogenicity analysis in the G. mellonella infection model supported the above conclusion that larvae infected with T6SS-positive hvKp isolates showed a worse survival rate than the other groups (p < 0.05). Alternately, we found that the T6SS-negative hvKp isolate group had a worse survival rate than the T6SS-positive cKp isolates. This result might indicate that virulence genes were a larger contributor to survival rate than T6SS genes. However, this remains speculative, and no direct experimental results support this notion.
T6SS-positive isolates had stronger biofilm-forming abilities than T6SS-negative isolates. This was in agreement with the findings of Liao et al. but in contrast with Zhang et al. (2022) and Liao et al. (2022). T6SS genes are related to biofilm formation, but the role of T6SS has been controversial, and its specific mechanism needs further study (Chen et al., 2020; Dadashi et al., 2021; Fei et al., 2022).
In conclusion, this investigation showed the high virulence potential of T6SS-positive K. pneumoniae isolates, and T6SS genes may be involved in the hvKp virulence mechanism. The T6SS-positive isolates were more likely to be acquired from community infections. Most of the dominant virulence-associated factors (hvKp, K1, ST23, rmpA, rmpA2, and aerobactin) were highly clustered in T6SS-positive K. pneumoniae isolates and displayed lower rates of antimicrobial resistance, higher virulence in the in vivo G. mellonella infection model, and stronger biofilm abilities than T6SS-negative isolates. The limitations of this study are as follows: This was a single-center study; therefore, the molecular and clinical characteristics of K. pneumoniae isolates was regional. Multi-center studies would improve data reliability and further confirm our findings. Our future studies will aim to study the specific mechanism underlying how T6SS gene affects the virulence of K. pneumoniae.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Author contributions
WL proposed and directed the study. PL, AY, and BT performed the experiments and collected the clinical data and prepared the first draft. AY and ZW analyzed the data. WL, PL, AY, BT, ZW, ZJ, YL, JW, BZ, and QY revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Natural Science Foundation of China (grant number 81672066) and National Key R&D Program of China (No. 2018YFC2000203).
Acknowledgments
We thank all the staff in the Microbiology Department of Xiangya Hospital for their kind help.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1181701/full#supplementary-material
Footnotes
References
Altayb, H. N., Elbadawi, H. S., Baothman, O., Kazmi, I., Alzahrani, F. A., Nadeem, M. S., et al. (2022). Genomic analysis of multidrug-resistant Hypervirulent (Hypermucoviscous) Klebsiella pneumoniae strain lacking the Hypermucoviscous regulators (rmpA/rmpA2). Antibiotics (Basel) 11:596. doi: 10.3390/antibiotics11050596
Anderson, D. J., Chen, L. F., Weber, D. J., Moehring, R. W., Lewis, S. S., Triplett, P. F., et al. (2017). Enhanced terminal room disinfection and acquisition and infection caused by multidrug-resistant organisms and Clostridium difficile (the benefits of enhanced terminal room disinfection study): a cluster-randomised, multicentre, crossover study. Lancet 389, 805–814. doi: 10.1016/S0140-6736(16)31588-4
Barbosa, V. A. A., and Lery, L. M. S. (2019). Insights into Klebsiella pneumoniae type VI secretion system transcriptional regulation. BMC Genomics 20:506. doi: 10.1186/s12864-019-5885-9
Chen, L., Zou, Y., Kronfl, A. A., and Wu, Y. (2020). Type VI secretion system of Pseudomonas aeruginosa is associated with biofilm formation but not environmental adaptation. Microbiology 9:e991. doi: 10.1002/mbo3.991
Choby, J. E., Howard-Anderson, J., and Weiss, D. S. (2020). Hypervirulent Klebsiella pneumoniae -clinical and molecular perspectives. J. Intern. Med. 287, 283–300. doi: 10.1111/joim.13007
Cienfuegos-Gallet, A. V., Zhou, Y., Ai, W., Kreiswirth, B. N., Yu, F., and Chen, L. (2022). Multicenter genomic analysis of Carbapenem-resistant Klebsiella pneumoniae from bacteremia in China. Microbiol. Spectr. 10:e0229021. doi: 10.1128/spectrum.02290-21
Dadashi, M., Chen, L., Nasimian, A., Ghavami, S., and Duan, K. (2021). Putative RNA ligase RtcB affects the switch between T6SS and T3SS in Pseudomonas aeruginosa. Int. J. Mol. Sci. 22:12561. doi: 10.3390/ijms222212561
Dong, N., Liu, L., Zhang, R., Chen, K., Xie, M., Chan, E. W. C., et al. (2019). An IncR plasmid harbored by a Hypervirulent Carbapenem-resistant Klebsiella pneumoniae strain possesses five tandem repeats of the Bla(KPC-2)::NTE(KPC)-id fragment. Antimicrob. Agents Chemother. 63, e01775–e01718. doi: 10.1128/AAC.01775-18
Du, P., Liu, C., Fan, S., Baker, S., and Guo, J. (2022). The role of plasmid and resistance Gene Acquisition in the emergence of ST23 multi-drug resistant. Microbiol. Spectr. 10:e0192921. doi: 10.1128/spectrum.01929-21
Effah, C. Y., Sun, T., Liu, S., and Wu, Y. (2020). Klebsiella pneumoniae: an increasing threat to public health. Ann. Clin. Microbiol. Antimicrob. 19:1. doi: 10.1186/s12941-019-0343-8
Fan, L. P., Yu, Y., Huang, S., Liao, W., Huang, Q. S., Du, F. L., et al. (2022). Genetic characterization and passage instability of a novel hybrid virulence plasmid in a ST23 hypervirulent Klebsiella pneumoniae. Front. Cell. Infect. Microbiol. 12:870779. doi: 10.3389/fcimb.2022.870779
Fei, N., Ji, W., Yang, L., Yu, C., Qiao, P., Yan, J., et al. (2022). Hcp of the type VI secretion system (T6SS) in Acidovorax citrulli group II strain Aac5 has a dual role as a Core structural protein and an effector protein in colonization, growth ability, competition, biofilm formation, and ferric Iron absorption. Int. J. Mol. Sci. 23:9632. doi: 10.3390/ijms23179632
Gu, D., Dong, N., Zheng, Z., Lin, D., Huang, M., Wang, L., et al. (2018). A fatal outbreak of ST11 carbapenem-resistant hypervirulent Klebsiella pneumoniae in a Chinese hospital: a molecular epidemiological study. Lancet Infect. Dis. 18, 37–46. doi: 10.1016/S1473-3099(17)30489-9
Hernandez, M., Lopez-Urrutia, L., Abad, D., De Frutos Serna, M., Ocampo-Sosa, A. A., and Eiros, J. M. (2021). First report of an extensively drug-resistant ST23 Klebsiella pneumoniae of capsular serotype K1 co-producing CTX-M-15, OXA-48 and ArmA in Spain. Antibiotics (Basel) 10:157. doi: 10.3390/antibiotics10020157
Hsieh, P. F., Lu, Y. R., Lin, T. L., Lai, L. Y., and Wang, J. T. (2019). Klebsiella pneumoniae type VI secretion system contributes to bacterial competition, cell invasion, Type-1 fimbriae expression, and in vivo colonization. J. Infect. Dis. 219, 637–647. doi: 10.1093/infdis/jiy534
Hui, X., Chen, Z., Zhang, J., Lu, M., Cai, X., Deng, Y., et al. (2021). Computational prediction of secreted proteins in gram-negative bacteria. Comput. Struct. Biotechnol. J. 19, 1806–1828. doi: 10.1016/j.csbj.2021.03.019
Kamau, E., Ranson, E. L., Tsan, A. T., Bergmann-Leitner, E. S., Garner, O. B., and Yang, S. (2022). Clinical and genomic characterization of hypervirulent Klebsiella pneumoniae (hvKp) infections via passive surveillance in Southern California, 2020-2022. Front. Microbiol. 13:1001169. doi: 10.3389/fmicb.2022.1001169
Lai, C. C., and Yu, W. L. (2021). Klebsiella pneumoniae harboring Carbapenemase genes in Taiwan: its evolution over 20 years, 1998-2019. Int. J. Antimicrob. Agents 58:106354. doi: 10.1016/j.ijantimicag.2021.106354
Lam, M. M. C., Wick, R. R., Watts, S. C., Cerdeira, L. T., Wyres, K. L., and Holt, K. E. (2021). A genomic surveillance framework and genotyping tool for Klebsiella pneumoniae and its related species complex. Nat. Commun. 12:4188. doi: 10.1038/s41467-021-24448-3
Lee, C. R., Lee, J. H., Park, K. S., Jeon, J. H., Kim, Y. B., Cha, C. J., et al. (2017). Antimicrobial resistance of Hypervirulent Klebsiella pneumoniae: epidemiology, Hypervirulence-associated determinants, and resistance mechanisms. Front. Cell. Infect. Microbiol. 7:483. doi: 10.3389/fcimb.2017.00483
Lewis, M. P. W. J. (2022). Clinical and laboratory standards institute (CLSI). Performance stands for antimicrobial susceptibility testing 2022:the M100-S130.
Liao, W., Huang, H. H., Huang, Q. S., Fang-Ling, D., Dan Wei, D., La-Gen, W., et al. (2022). Distribution of type VI secretion system (T6SS) in clinical Klebsiella pneumoniae strains from a Chinese hospital and its potential relationship with virulence and drug resistance. Microb. Pathog. 162:105085. doi: 10.1016/j.micpath.2021.105085
Lin, J., Huang, Y., Qian, L., Pan, X., and Song, Y. (2022). Liver abscess combined with endogenous Endophthalmitis caused by genotype ST25 serotype K2 Hypervirulent Klebsiella pneumoniae: a case report. Infect Drug Resist. 15, 4557–4561. doi: 10.2147/IDR.S376443
Liu, S., Ding, Y., Xu, Y., Li, Z., Zeng, Z., and Liu, J. (2022). An outbreak of extensively drug-resistant and hypervirulent Klebsiella pneumoniae in an intensive care unit of a teaching hospital in Southwest China. Front. Cell. Infect. Microbiol. 12:979219. doi: 10.3389/fcimb.2022.979219
Liu, C., and Guo, J. (2018). Characteristics of ventilator-associated pneumonia due to hypervirulent Klebsiella pneumoniae genotype in genetic background for the elderly in two tertiary hospitals in China. Antimicrob. Resist. Infect. Control 7:95. doi: 10.1186/s13756-018-0371-8
Liu, C., and Guo, J. (2019). Hypervirulent Klebsiella pneumoniae (hypermucoviscous and aerobactin positive) infection over 6 years in the elderly in China: antimicrobial resistance patterns, molecular epidemiology and risk factor. Ann. Clin. Microbiol. Antimicrob. 18:4. doi: 10.1186/s12941-018-0302-9
Liu, Y. M., Li, B. B., Zhang, Y. Y., Zhang, W., Shen, H., Li, H., et al. (2014). Clinical and molecular characteristics of emerging hypervirulent Klebsiella pneumoniae bloodstream infections in mainland China. Antimicrob. Agents Chemother. 58, 5379–5385. doi: 10.1128/AAC.02523-14
Mei, Y. F., Liu, P. P., Wan, L. G., Liu, Y., Wang, L. H., Wei, D. D., et al. (2017). Virulence and genomic feature of a virulent Klebsiella pneumoniae sequence type 14 strain of serotype K2 harboring Bla(NDM-5) in China. Front. Microbiol. 8:335. doi: 10.3389/fmicb.2017.00335
Morra, R., Del Carratore, F., Muhamadali, H., Horga, L. G., Halliwell, S., Goodacre, R., et al. (2018). Translation stress positively regulates MscL-dependent excretion of cytoplasmic proteins. MBio 9, e02118–e02117. doi: 10.1128/mBio.02118-17
Nicolo, S., Mattiuz, G., Antonelli, A., Arena, F., Di Pilato, V., Giani, T., et al. (2022). Hypervirulent Klebsiella pneumoniae strains modulate human dendritic cell functions and affect T(H)1/T(H)17 response. Microorganisms 10:384. doi: 10.3390/microorganisms10020384
Russo, T. A., and Gulick, A. M. (2019). Aerobactin synthesis proteins as Antivirulence targets in Hypervirulent Klebsiella pneumoniae. ACS Infect Dis 5, 1052–1054. doi: 10.1021/acsinfecdis.9b00117
Sohrabi, M., Alizade Naini, M., Rasekhi, A., Oloomi, M., Moradhaseli, F., Ayoub, A., et al. (2022). Emergence of K1 ST23 and K2 ST65 hypervirulent klebsiella pneumoniae as true pathogens with specific virulence genes in cryptogenic pyogenic liver abscesses shiraz Iran. Front. Cell. Infect. Microbiol. 12:964290. doi: 10.3389/fcimb.2022.964290
Su, S., Zhang, J., Zhao, Y., Yu, L., Wang, Y., Wang, Y., et al. (2020). Outbreak of KPC-2 Carbapenem-resistant Klebsiella pneumoniae ST76 and Carbapenem-resistant K2 Hypervirulent Klebsiella pneumoniae ST375 strains in Northeast China: molecular and virulent characteristics. BMC Infect. Dis. 20:472. doi: 10.1186/s12879-020-05143-y
Suay-Garcia, B., and Perez-Gracia, M. T. (2019). Present and future of Carbapenem-resistant Enterobacteriaceae (CRE) infections. Antibiotics (Basel) 8:122. doi: 10.3390/antibiotics8030122
Walker, K. A., Treat, L. P., Sepulveda, V. E., and Miller, V. L. (2020). The small protein RmpD drives Hypermucoviscosity in Klebsiella pneumoniae. MBio 11, e01750–e01720. doi: 10.1128/mBio.01750-20
Wang, Z., Guo, G., Li, Q., Li, P., Li, M., Zhou, L., et al. (2022). Combing Immunoinformatics with Pangenome analysis to design a multiepitope subunit vaccine against Klebsiella pneumoniae K1, K2, K47, and K64. Microbiol. Spectr. 10:e0114822. doi: 10.1128/spectrum.01148-22
Wick, R. R., Judd, L. M., Gorrie, C. L., and Holt, K. E. (2017). Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 13:e1005595. doi: 10.1371/journal.pcbi.1005595
Wyres, K. L., Lam, M. M. C., and Holt, K. E. (2020). Population genomics of Klebsiella pneumoniae. Nat. Rev. Microbiol. 18, 344–359. doi: 10.1038/s41579-019-0315-1
Xu, T., Wu, X., Cao, H., Pei, T., Zhou, Y., Yang, Y., et al. (2022). The characteristics of multilocus sequence typing, virulence genes and drug resistance of Klebsiella pneumoniae isolated from cattle in northern Jiangsu, China. Animals (Basel) 12:2627. doi: 10.3390/ani12192627
Yu, K. W., Xue, P., Fu, Y., and Yang, L. (2021). T6SS mediated stress responses for bacterial environmental survival and host adaptation. Int. J. Mol. Sci. 22:478. doi: 10.3390/ijms22020478
Zhang, Y., Xu, Y., and Huang, Y. (2022). Virulence genotype and correlation of clinical Severeness with presence of the type VI secretion system in Klebsiella pneumoniae isolates causing bloodstream infections. Infect Drug Resist 15, 1487–1497. doi: 10.2147/IDR.S353858
Zhang, Y., Zhao, C., Wang, Q., Wang, X., Chen, H., Li, H., et al. (2016). High prevalence of Hypervirulent Klebsiella pneumoniae infection in China: geographic distribution, clinical characteristics, and antimicrobial resistance. Antimicrob. Agents Chemother. 60, 6115–6120. doi: 10.1128/AAC.01127-16
Zhao, B., Hu, R., Gong, L., Wang, X., Zhu, Y., and Wu, G. (2021). Pyogenic liver abscess and endogenous Endophthalmitis due to K64-ST1764 Hypervirulent Klebsiella pneumoniae: a case report. Infect Drug Resist 14, 71–77. doi: 10.2147/IDR.S289088
Zheng, J. X., Lin, Z. W., Chen, C., Chen, Z., Lin, F. J., Wu, Y., et al. (2018). Biofilm formation in Klebsiella pneumoniae bacteremia strains was found to be associated with CC23 and the presence of wcaG. Front. Cell. Infect. Microbiol. 8:21. doi: 10.3389/fcimb.2018.00021
Zhou, M., Lan, Y., Wang, S., Liu, Q., Jian, Z., Li, Y., et al. (2020). Epidemiology and molecular characteristics of the type VI secretion system in Klebsiella pneumoniae isolated from bloodstream infections. J. Clin. Lab. Anal. 34:e23459. doi: 10.1002/jcla.23459
Keywords: type VI system (T6SS), hypervirulent Klebsiella pneumoniae (hvKP), abscess, hypervirulent, virulence
Citation: Liu P, Yang A, Tang B, Wang Z, Jian Z, Liu Y, Wang J, Zhong B, Yan Q and Liu W (2023) Molecular epidemiology and clinical characteristics of the type VI secretion system in Klebsiella pneumoniae causing abscesses. Front. Microbiol. 14:1181701. doi: 10.3389/fmicb.2023.1181701
Edited by:
Juan A. Ayala, Autonomous University of Madrid, SpainReviewed by:
Hussein O. M. Al-Dahmoshi, University of Babylon, IraqShuwei Zheng, Sengkang General Hospital, Singapore
Copyright © 2023 Liu, Yang, Tang, Wang, Jian, Liu, Wang, Zhong, Yan and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wenen Liu, wenenliu@163.com
†These authors have contributed equally to this work
 Peilin Liu
Peilin Liu Awen Yang1†
Awen Yang1† Bin Tang
Bin Tang Zhiqian Wang
Zhiqian Wang Yanjun Liu
Yanjun Liu Wenen Liu
Wenen Liu