- 1Key Laboratory of Environmental Pollution Monitoring and Disease Control, Ministry of Education, Guizhou Medical University, Guiyang, China
- 2School of Public Health, Guizhou Medical University, Guiyang, China
- 3Guizhou Provincial Engineering Research Center of Food Nutrition and Health, Guiyang, China
- 4Shenzhen Key Laboratory of Marine Biotechnology and Ecology, College of Life Science and Oceanography, Shenzhen University, Shenzhen, China
- 5Key Laboratory of Optimal Utilization of Natural Medicine Resources, School of Pharmaceutical Sciences, Guizhou Medical University, Guiyang, China
- 6State Key Laboratory of Functions and Applications of Medicinal Plants, Guizhou Medical University, Guiyang, China
The purpose of this study is to understand the mechanism of sodium arsenite (NaAsO2)-induced apoptosis of L-02 human hepatic cells, and how Dictyophora polysaccharide (DIP) protects L-02 cells from arsenic-induced apoptosis. The results revealed that DIP pretreatment inhibited NaAsO2 induced L-02 cells apoptosis by increasing anti-apoptotic Bcl-2 expression and decreasing pro-apoptotic Bax expression. Proteomic analysis showed that arsenic treatment disrupted the expression of metabolism and apoptosis associated proteins, including ribosomal proteins (RPs). After pretreatment with DIP, the expression levels of these proteins were reversed or restored. For the first time, it was observed that the significant decrease of cytoplasmic RPs and the increase of mitochondrial RPs were related to human normal cell apoptosis induced by arsenic. This is also the first report that the protective effect of DIP on cells was related to RPs. The results highlight the relationship between RPs and apoptosis, as well as the relationship between RPs and DIP attenuating arsenic-induced apoptosis.
Introduction
Arsenic is known as a serious environmental toxin and human carcinogen (Bhattacharya et al., 2014). According to global epidemiological statistics, more than 200 million people currently suffer from arsenic exposure, particularly in developing countries (Adil et al., 2016). Arsenic exposure caused human health problems involve multiple tissues and organs, especially in the cardiovascular system and liver (Goudarzi et al., 2018). Environmental exposure to arsenic is undoubtedly now a major global public health problem, as well as a serious social and medical problem (Chen et al., 2012; Wang et al., 2020a). Sodium arsenite (NaAsO2) is the most toxic substance of different arsenic compounds in our living environment (Duan et al., 2020; Lv et al., 2020). Accumulation of arsenic in the human body can lead to organ damage and tissue canceration. In particular, the liver is one of the target organs of arsenic in humans (Dash et al., 2018; Lv et al., 2020). The cellular and molecular biological experiment has shown that arsenic increases the production of reactive oxygen species (ROS) by inhibiting the activity of antioxidant enzymes. Arsenic can also cause protein oxidation, DNA damage, and apoptosis (Lin et al., 2008; Adil et al., 2015; Goudarzi et al., 2018). However, there is still no broad consensus about the exact mechanism of arsenic-induced toxicity.
Currently, treatments for arsenic poisoning include hemodialysis or chemotherapy, using chelating agents and adsorbents. However, elevated blood pressure and other toxic effects may occur (Saha et al., 2016). Bioactive natural compounds commonly found in dietary plants can treat arsenic-induced toxicity, attracting increased attention to their little or no side effects (Ola-Davies and Akinrinde, 2016; Goudarzi et al., 2018; Perker et al., 2019), and more attention has been paid to their medicinal value (Kanwal et al., 2018; Habtemariam, 2019). Among them, Dictyophora has been reported to act on anti-inflammatory, antioxidant, hypoglycemic, and lipid-lowering effects (Zhang et al., 2016; Han et al., 2017; Wang Y. et al., 2019). Furthermore, the polysaccharide extracted from Dictyophora has a protective effect on the liver (Wang et al., 2019a; Wang et al., 2019b; Hu et al., 2020). However, the effect of Dictyophora polysaccharide (DIP) on sodium arsenite induced hepatotoxicity is still unclear.
In this study, DIP was shown to protect human normal liver cells L-02 from the sodium arsenic induced toxicity, and a comparative proteomics analysis based on iTRAQ (isobaric tags for relative and absolute quantification) was performed to explore the molecular mechanism of arsenic-induced apoptosis and the protective effect of DIP on arsenic-induced hepatotoxicity.
Materials and Methods
Chemicals
Food-grade Dictyophora was provided by Zhijin Sifang Hongye (Zhijin City, Guizhou Province, China). Sodium arsenite (NaAsO2) was obtained from Sigma Chemical Corp (St. Louis, MO, United States). Human normal hepatocytes (L-02 cells) were purchased from the Shanghai Cell Bank of the Chinese Academy of Sciences (Shanghai, China). RPMI 1640 cell culture medium, trypsin, and fetal bovine serum (FBS) were purchased from Gibco Company (California, United States). Phosphate buffer saline (PBS) was purchased from Zhongsha Jinqiao Biotechnology Co., Ltd. (Beijing, China). Dimethyl sulfoxide (DMSO) was obtained from Sigma (St. Louis, MO, United States). The BCA protein detection kit, protein sample buffer, and Western blot analysis gel preparation kit were purchased from Beyotime Biotechnology Co., Ltd. (Shanghai, China). Protein molecular weight markers were obtained from Fermentas (Burlington, Canada). Polyvinylidene fluoride (PVDF) film and enhanced chemiluminescence (ECL) kit were purchased from Bio-Rad (California, United States). Cell Counting Kit-8 (CCK-8), RIPA lysis buffer, rabbit anti-human Bcl-2, Bax, β-actin, GAPDH, ribosomal protein S5 (Rps5), and 14-3-3 protein sigma (SFN) antibodies and horseradish peroxidase (HRP) labeled secondary antibodies were purchased from Boster Biological Technology, Ltd. (Boster, Wuhan, China).
Dictyophora Polysaccharide Preparation
The separation and purification of DIP was performed as previously described with minor modifications (Liao et al., 2015; Zhang et al., 2016; Yu et al., 2017; Hu et al., 2020). Briefly, the fruit body of Dictyophora was dried at 45°C for 2 h, ground into powder, extracted in boiling water for 3 h at a water-to-material ratio (1:20), and then centrifuged at 4,000 × g for 15 min. The supernatant was extracted and concentrated at 50°C using a rotary evaporator (R-215, Buchi, Switzerland). The concentrated extract was collected and proteins were removed using the Sevage method. The crude polysaccharides of Dictyophora were obtained by freeze-drying after overnight precipitation with 4 volumes of 100% ethanol. The content of sugar was determined by using phenol-sulfuric acid colorimetric assay. The content was 84.13%. According to our experimental data, 10 mg DIP can be obtained from 80 mg of dried Dictyophora powder.
Preparation of Dictyophora Polysaccharide Stock Solution
The dry product of 400 mg DIP was dissolved in 20 ml serum-free DMEM medium to prepare a final concentration of 20 mg/ml mother liquor, filtered and stored at −20°C. When in use, it is diluted to the corresponding concentration with serum-free DMEM medium.
Cell Culture
L-02 cells were cultured in a 5% CO2 incubator at 37°C. The control group used DMEM high glucose medium containing 10% FBS and 1% penicillin/streptomycin, and the treatment group was pretreated with NaAsO2 or DIP for 4 h and then exposed to NaAsO2. All experiments were performed 24 h after cell inoculation.
Cell Counting Kit-8 Assay
The cell viability of L-02 cells was measured by CCK-8 assay. Cells were seeded in 96-well plates at a density of 1 × 104 cells per well, and were treated with different concentration of NaAsO2 for 24 h. In order to explore the intervention effect of DIP, L-02 cells were pretreated with different concentration DIP solution for 4 h, and then treated with of 10 μM NaAsO2 for 24 h. In addition, the viability of cells pretreated with DIP (80 μg/ml) and then exposed to different concentrations sodium arsenate was also investigated. CCK-8 reagent was added to each well and incubated at 37°C for 2 h according to the manufacturer protocol. Microplate Reader (Thermo Fisher Scientific) was used to measure the absorbance at 450 nm to determine cell viability.
Annexin V/PI Assay
The percentage of apoptotic cells was detected by Annexin V-FITC apoptosis detection kit (Beyotime, Shanghai, China). After NaAsO2 (10 μM) treatment or DIP (80 μg/ml) pretreatment followed by exposure to NaAsO2, the L-02 cells were collected and washed with PBS, and re-suspended by adding 100 μL binding buffer, and then incubated with 5 µL Annexin V-FITC, and 10 µL PI at room temperature. The apoptosis rate was detected by flow cytometry (ACEA NovoCyte, United States).
Protein Extraction for Proteomics Analysis
After treatment with NaAsO2 (10 μM) or DIP (80 μg/ml) pretreatment and then exposed to 10 μM NaAsO2, L-02 cells were rinsed twice with ice-cold PBS. The cells were harvested and resuspended in lysis buffer (8 M urea, 2 mM EDTA, 10 mM DTT, and 1% protease inhibitor cocktail). After sonicated and centrifuged at 13,000 g at 4°C for 10 min to remove debris, the protein in supernatant was precipitated with cold acetone for 2 h at −20°C. After centrifugation at 4°C at 12,000 × g for 10 min, the protein deposit was redissolved by urea buffer [8 M urea, 100 mM TEAB (triethylammonium bicarbonate)]. The protein concentration was detected using Bradford protein assay kit (Beyotime).
Trypsin Digestion
For trypsin digestion, 100 μg protein of each sample was first reduced with 10 mM DTT at 37°C for 60 min and then alkylated with 55 mM iodoacetamide (IAM) at room temperature for 30 min in darkness. The urea content of protein extract was diluted by adding 100 mM TEAB less than 2 M. The protein pool of each sample was digested with trypsin with the ratio of protein: trypsin = 50:1 mass ratio at 37°C overnight and 100:1 for a second digestion at 4 h.
Peptides Isobaric Tags for Relative and Absolute Quantification Labeling
After trypsin digestion, the peptides were desalted by Strata X SPE column and vacuum-dried. The digested peptides were then labeled with the iTRAQ reagents (AB Sciex), as follows: the control group was labeled with iTRAQ 113 and 114, while NaAsO2 treatment group with iTRAQ 115 and 116, and DIP pretreatment and NaAsO2 treatment group with iTRAQ 117 and 118. Briefly, peptides were reconstituted in 20 μl 500 mM TEAB and processed according to the manufacturer’s protocol for 8-plex iTRAQ kit (AB Sciex, Foster City, CA, United States). One unit of iTRAQ reagent was applied to the peptide solution after thawed and dissolved in 50 μL isopropanol. The peptide mixtures were incubated for 2 h at room temperature, then pooled and dried by vacuum centrifugation.
High-Performance Liquid Chromatographic Fractionation
The dried and labeled peptide was reconstituted with HPLC solution A [2% ACN (acetonitrile), pH 10] and then fractionated into high pH reverse-phase HPLC fractions using Waters Bridge Peptide BEH C18 (130 Å, 3.5 μm, 4.6 × 250 mm). Peptides were first separated by a gradient of 2–98% acetonitrile in pH 10 at a speed of 0.6 ml/min over 88 min into 48 fractions. The peptides were then mixed into 15 fractions and dried by vacuum centrifugation. The peptide fractions were desalted using Ziptip C18 (Millipore, MA, United States). Samples were finally dried under vacuum and kept at −20°C until they were analyzed by MS (mass spectrometry).
High-Resolution LC-MS/MS Analysis
The experiment was then performed by NanoLC 1000 LC-MS/MS using a Proxeon EAsY-nLC 1000 coupled to Q-Exactive mass spectrometer (Thermo Fisher Scientific, United States). Trypsin digestion fractions were reconstituted in 0.1% FA (formic acid) and immediately charged to the reversed-phase pre-column (Acclaim PepMap®100 C18, 3 μm, 100 Å, 75 μm × 2 cm) at 5 μl/min in 100% solvent A (0.1 M acetic acid in water). Next, peptides eluted from the trap column were loaded into a reversed-phase analytical column (Acclaim PepMap® RSLC C18, 2 μm, 100 Å, 50 μm × 15 cm). The gradient was comprised of an increase from 0 to 8% solvent B 0.1% FA in 98% ACN over 5 min, 8–25% solvent B over 35 min, 25–98% solvent B during 10 min and keep in 98% in 8 min at a constant flow rate of 300 nL/min at EAsY-nLC 1000 system. The eluent was sprayed from an NSI source at an electrospray voltage of 2.5 kV and then analyzed by tandem mass spectrometry (MS/MS) in Q Exactive. The mass spectrometer was operated in data-dependent mode, automatically switching between MS and MS/MS. Full-scan MS spectra (from m/z 300–2000) were acquired in the Orbitrap with a resolution of 70,000. Ion fragments were detected in the Orbitrap at a resolution of 17,500. The 15 most intense precursors were selected for subsequent decision tree-based ion trap HCD fragmentation at the collision energy of 32% in the MS survey scan with 10.0 s dynamic exclusion.
Data Processing and Isobaric Tags for Relative and Absolute Quantification
The resulting MS/MS raw data was searched against the transcriptome database using Sequest software integration in Proteome Discoverer (version 1.3, Thermo Scientific). The quest parameters were as follows: trypsin as a digestion enzyme, two missing cleavages, oxidized methionine, acetylation in N-Term, iTRAQ modification at the N-terminus of the peptide and iTRAQ 8-plex (K, Y) as the variable modification, fixed modifications like carbamidomethyl (C). The peptide mass tolerance and fragment mass tolerance were set to 20 ppm and 0.05 Da, respectively. A decoy database search strategy was adopted to estimate the false discovery rate (FDR) for peptide identification. For this study, a high peptide confidence (1% FDR) was selected. The cut-off values of 1.2-fold for up-regulated and 0.83-fold for down-regulated proteins, p-value < 0.05, were set as differentially expressed proteins (DEPs) (Shen et al., 2021).
Bioinformatics Analysis
Bioinformatics analysis was performed by using OMICSBEAN online tools (http://www.omicsbean.cn/) and String (Search Tool for the Retrieval of Interacting Genes/Proteins, version 9.1, http://string-db.org/) database. DEPs were analyzed by GO (gene ontology) annotation, KEGG (Kyoto Encyclopedia of Genes and Genomes) pathways, and protein-protein interaction (PPI) networks (Iqbal et al., 2018; Wei et al., 2018). Functional interaction network analysis was conducted using the ClueGO Cytoscape plugin (Bindea et al., 2009). The GO categories and pathways searched include biological processes (BP), cellular components (CC), molecular function (MF), KEGG (Kyoto Encyclopedia of Genes and Genomes), REACTOME, and Wiki pathway.
Western Blot Analysis
L-02 cells were incubated in 6-well plates. After being treated with NaAsO2 or DIP as indicated, cells were collected and lysed with RIPA buffer containing 1 mM PMSF and 1% protease inhibitor cocktail. The protein concentration was measured using the BCA kit. After SDS-PAGE electrophoresis transformation, the separated proteins were transferred to a polyvinylidene fluoride (PVDF) membrane. The membrane was blocked with 5% skimmed milk at room temperature for 2 h and incubated with the indicated primary antibodies overnight at 4°C. After incubated with HRP-conjugated second antibody at room temperature for 1 h, the blotting signal was generated by chemiluminescence using an enhanced chemiluminescence (ECL) kit (Thermo Fisher Scientific). The gray value of protein band was analyzed by Image Labe software, and the ratio of target protein to internal reference gray value was used to reflect the expression of protein.
Statistical Analysis
Statistical analysis was carried out with SPSS Version 20.0 (SPSS Software, Chicago, IL, United States). The experimental data is shown as the means ± SD. Single factor analysis of variance (ANOVA) was used to detect the different distribution of various groups. The distribution of biometric values is normalized by logarithmic transformation. The statistically significant level was p < 0.05.
Results
Detection of Components of DIP by High-Performance Liquid Chromatographic
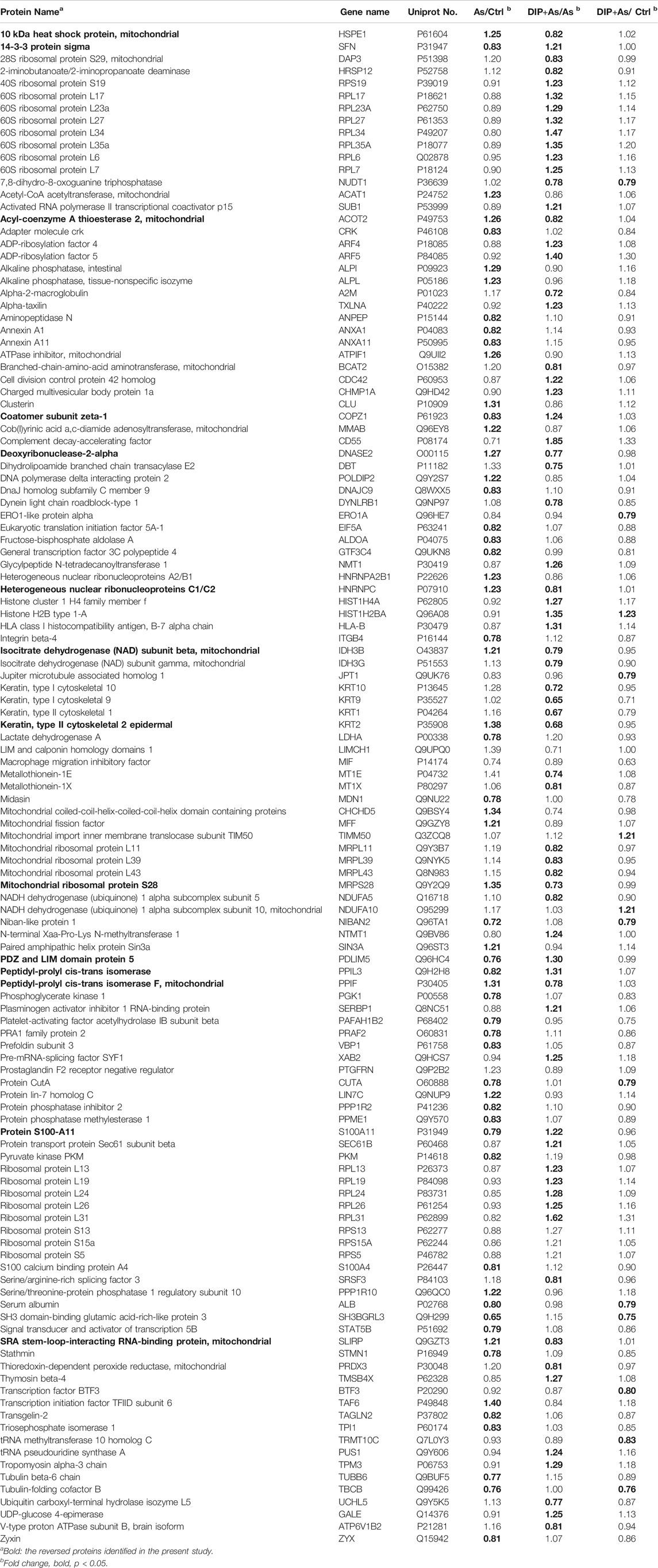
The HPLC detection showed that the monosaccharides in the Dictyophora polysaccharides (DIP) were D-mannose, D-glucose, D-galactose, D-xylose, L-fucose (Figure 1A), and their contents were 15.27, 523.57, 27.80, 10.01, and 17.84 mg/L respectively, of which D-glucose accounted for 88.07% (Supplementary Table S1).
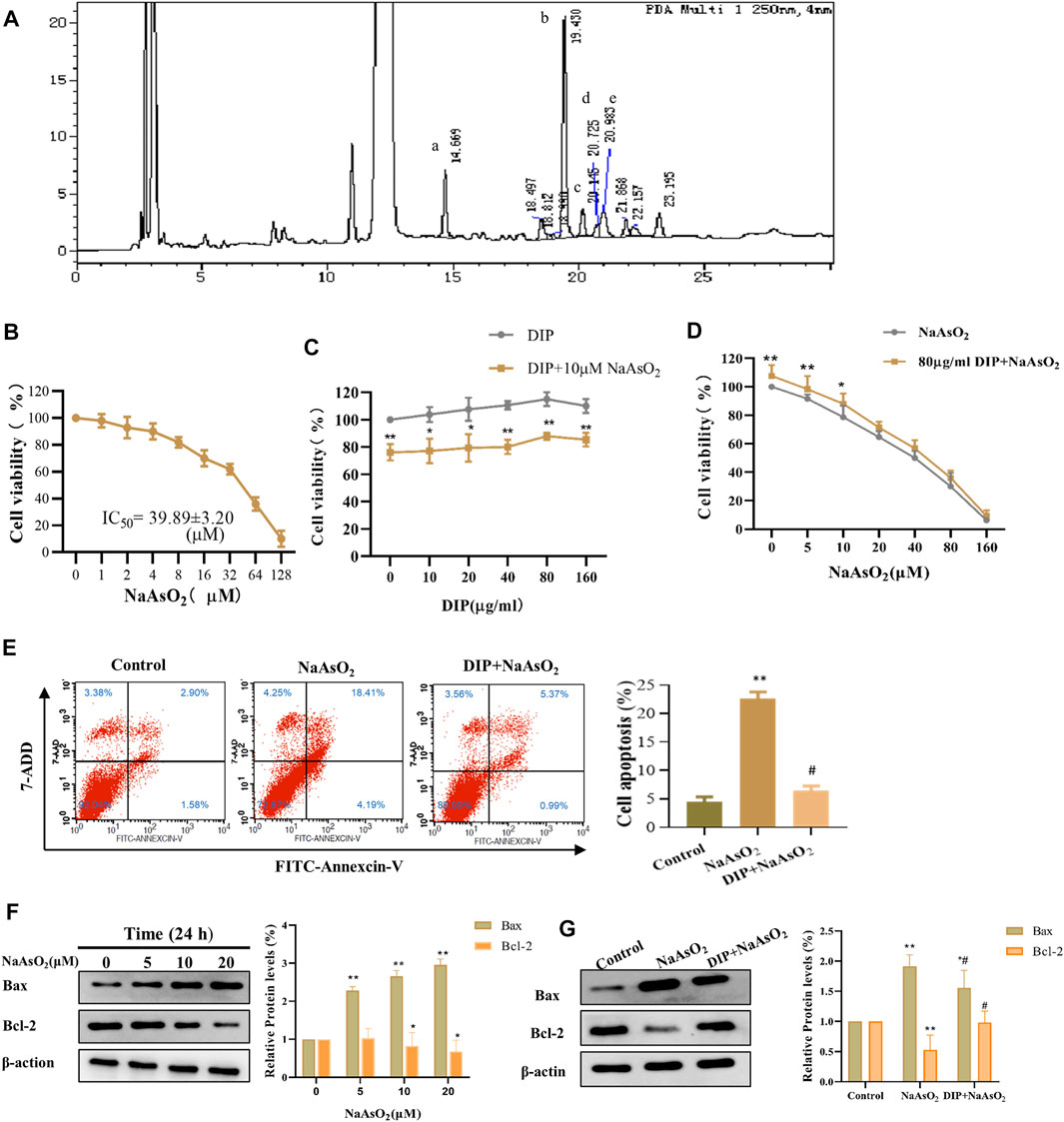
FIGURE 1. DIP against NaAsO2-induced hepatotoxicity in L-02 cells. (A) HPLC profile of sporocarp polysaccharide of Dictyophora. (a) D-mannose. (b) D-glucose. (c) D-galactose. (d) D-xylose. (e) L-fucose. (B) The viability of L-02 cells treated with different concentrations of NaAsO2 (CCK-8 assay). (C) The viability of L-02 cells exposed to arsenic (10 μM NaAsO2) with or without pretreatment with different concentrations of DIP. (D) The viability of L-02 cells pretreated with DIP (80 μg/ml) and then exposed to different concentrations of NaAsO2. (E) Flow cytometry profiles showing the distribution of cells (Left). Plot showing the proportion of apoptotic cells (Right). Compared with control group, **p < 0.01; compared with NaAsO2 group, #p < 0.05. (F) Western blot analysis of Bax or Bcl-2 protein expression levels when treated with different arsenic concentrations (Left). Plot showing Bax or Bcl-2 protein expression levels (Right). (G) Western blot analysis of Bax or Bcl-2 protein expression levels when pretreated with DIP and then treated with arsenic (Left). Plot showing Bax or Bcl-2 protein expression levels (Right). Compared with the control group, *p < 0.05, **p < 0.01. Compared with the control group, *p < 0.05, **p < 0.01; compared with NaAsO2 group, #p < 0.05.
DIP Antagonizes the Suppression Effect of NaAsO2 on L-02 Cell Viability
To investigate the cytotoxicity of sodium arsenite to L-02 cells, L-02 cells were treated with NaAsO2 treatment. We found that NaAsO2 reduced the viability of L-02 cells in a dose-dependent manner. The IC50 value was 39.89 ± 3.20 μM (Figure 1B). However, DIP pretreatment significantly improved L-02 cells viability, even when exposed to 1/8 of IC50 (5 μM) and 1/4 of IC50 (10 μM) NaAsO2. Pretreatment with 80 μg/ml DIP enabled L-02 cells to tolerate 10 μM sodium arsenite (Figures 1C,D). The results indicated that DIP has the potential to antagonize sodium arsenite cytotoxicity.
DIP Inhibits NaAsO2-Induced Apoptosis in L-02 Cells
Considering sodium arsenite is recognized as an apoptosis inducer (Bashir et al., 2006; Chen et al., 2012; Sun et al., 2017), we then investigated whether NaAsO2 could induce apoptosis in L-02 cells. Flow cytometry analysis showed that the proportion of apoptotic cells increased significantly after treatment with 10 μM NaAsO2 (Figure 1E). Furthermore, with the increasing dosage of NaAsO2 treatment, pro-apoptotic protein Bax was up-regulated, while anti-apoptotic Bcl-2 protein was down-regulated in L-02 cells (Figure 1F), suggesting NaAsO2 can induce the apoptosis of L-02 cells. However, DIP pretreatment not only significantly reduced the apoptosis induced by NaAsO2 (Figure 1E) but also reversed the expression of Bax and Bcl-2 (Figure 1G), revealing the protective role of DIP in resisting sodium arsenite-induced apoptosis of L-02 cells.
Overview of Protein Expression Characteristics of Different Groups
The iTRAQ analysis was further performed to explore the molecular mechanism of DIP against sodium arsenite-induced cytotoxicity in L-02 cells. We identified the protein expression profile in L-02 cells treated with 10 μM NaAsO2 compared with the control group (As/Ctrl group). After DIP pretreatment, the protein expression characteristics of arsenic treated group were also analyzed (DIP + As/As group). A total of 2,876 proteins were identified. Among them, 60, 71, and 13 proteins were identified as DEPs in As/Ctrl group, DIP + As/As group, and DIP + As/Ctrl groups (Figures 2A–C; Table 1), respectively. Of these, 14 DEPs were found to be common between the As/Ctrl group and DIP + As/As group, with the opposite expression trend in these two groups (named as reversed proteins; Figure 2D and Table 1). Cluster analysis showed that the protein expression characteristics of DIP + As/As group and DIP + As/Ctrl were more similar, but almost opposite to As/Ctrl group (Figure 2E).
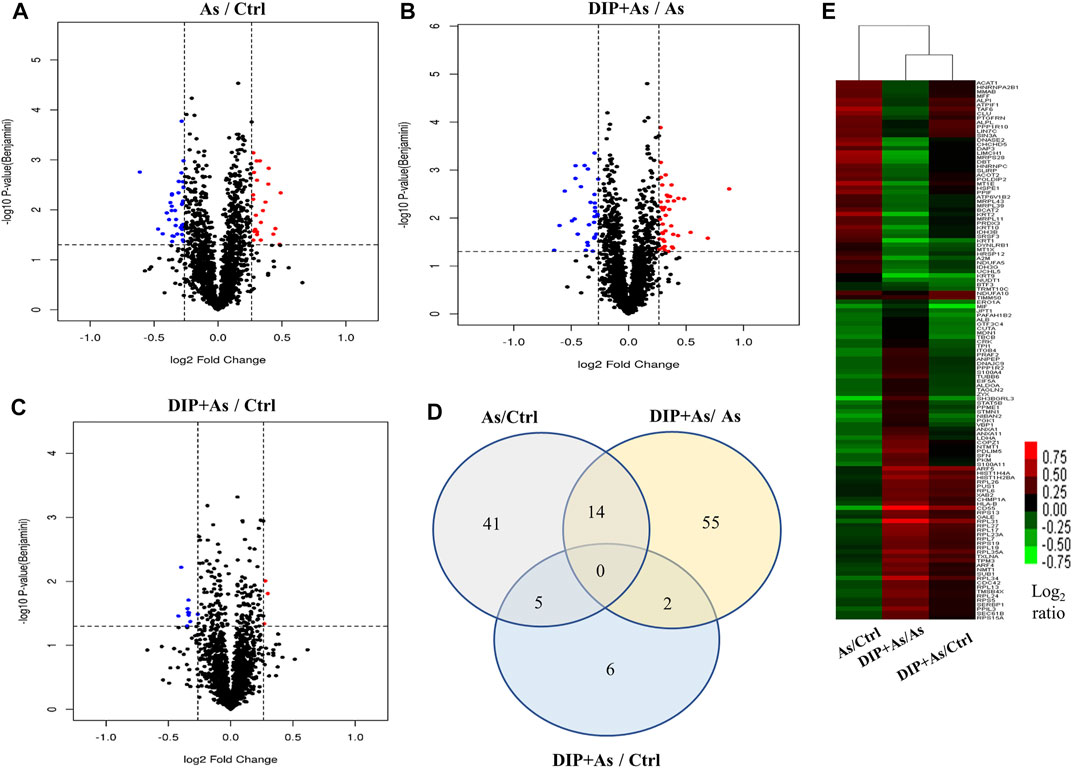
FIGURE 2. The differentially expressed proteins identified in different groups. (A) Volcano plots depicted the distribution of proteins in As/Ctrl group. (B) Volcano plots depicted the distribution of proteins in DIP + As/As group. (C) Volcano plots depicted the distribution of proteins in DIP + As/Ctrl group. (D) Venn diagrams of DEPs between the As/Ctrl group, DIP + As/As group, and DIP + As/Ctrl group. (E) Cluster map comparing the DEPs identified in As/Ctrl, DIP + As/As, and DIP + As/Ctrl groups. Red color indicates higher expression, green indicates lower expression, and black indicates similar expression levels. (A–C): The log2 fold change (FC) is plotted versus the –log10 of the p-value (Benjamini). Red dots = hits with p < 0.05 and mean log2FC > 0.83; blue dots = hits with p < 0.05 and means |log2FC| < 0.83.
Protein Expression Profile of NaAsO2-Treated L-02 Cells Compared With Natural Growth L-02 Cells (As/Ctrl Group).
By bioinformatics analysis, the BP, CC, MF, and KEGG pathways associated with these 60 DEPs in As/Ctrl group are presented in Figures 3A–D and Supplementary Table S2–5. The BP related to DEPs in As/Ctrl group involved mainly in the generation of precursor metabolites and energy, glycolytic process, ATP generation from ADP, pyruvate biosynthesis process, and NADH regeneration, etc. (Figure 3A). In particular, 14 DEPs were related to regulation of apoptotic process including ALB, ANXA1, ATPIF1, CLU, FAM129B, HSPE1, LDHA, MFF, MIF, PPIF, PPP1R10, SFN, SIN3A, and STAT5B (Supplemenntary Table S2). Four DEPs (CLU, MFF, PPIF, and SFN) were associated with apoptotic mitochondrial changes, 3 DEPs (MFF, POLDIP2, and SLIRP) were related to mitochondrion morphogenesis, and 3 DEPs (CLU, MFF, and SFN) were involved in the release of cytochrome c from mitochondria (Supplementary Table S2). The DEPs were mainly distributed in the cytoplasm, cytoplasmic parts, mitochondrion, and mitochondrial matrix, etc. (Figure 3B; Supplementary Table S3). The MF of these DEPs included protein binding, calcium-dependent protein binding, alkaline phosphatase activity, and protein phosphatase inhibitor activity, etc. (Supplementary Table S4).
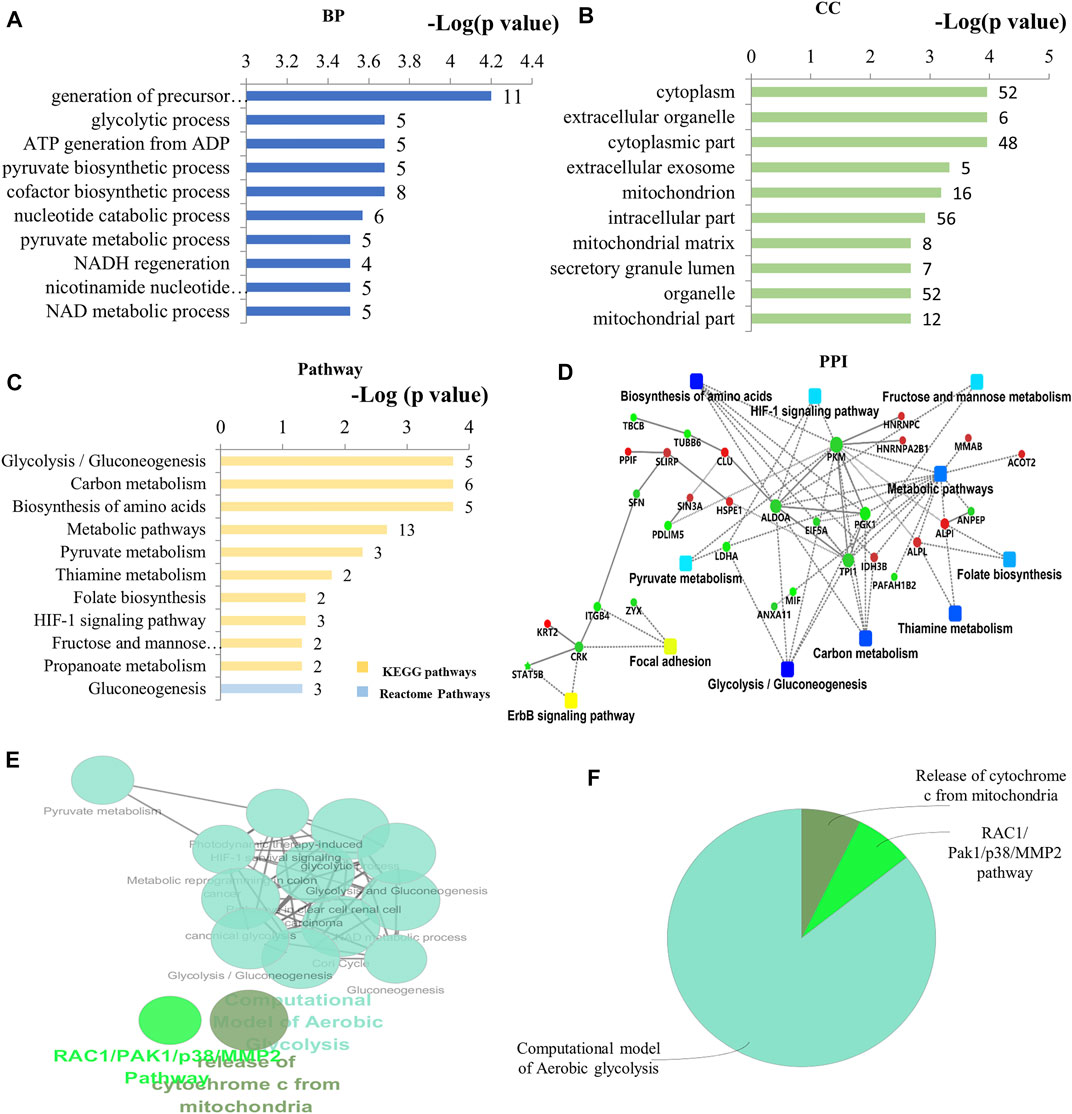
FIGURE 3. Bioinformatics analysis of the differentially expressed proteins in As/Ctrl group. (A) The top 10 ranking of BP associated with differentially expressed proteins. (B) The top 10 ranking of CC associated with differentially expressed proteins. (C) The significantly enriched pathways linked to differentially expressed proteins. (D) PPI network linked to the differentially expressed proteins in As/Ctrl group. Red: the expression of these proteins was up-regulated. Green: the expression of these proteins was down-regulated. (E) Functional interaction network analysis was performed by using ClueGO cytoscape plugin. (F) Overview the specific cluster. (A–C) Number of proteins associated with each category for gene-enrichment analysis is shown on the right of each term bar.
After NaAsO2 treatment, KEGG pathway analysis showed that most DEPs were involved in metabolic pathways, including glycolysis, carbon metabolism, pyruvate metabolism, amino acid biosynthesis and metabolic thiamine, folic acid biosynthesis, etc. (Figure 3C). Seven (7) DEPs were involved in these metabolism-related pathways, including ACAT1, ALDOA, IDH3B, LDHA, PGK1, PKM, and TPI1. Among them, 5 DEPs were down-regulated in L-02 cells, including ALDOA, LDHA, PGK1, PKM, TPI1 (Table 1; Supplementary Table S5). Other signal transduction pathways included the HIF-1 signaling pathway. The DEPs associated with this pathway include ALDOA, LDHA, and PGK1, which have also been down-regulated (Table 1; Supplementary Table S5). The PPI network associated with the DEPs is shown in Figure 3D. Consistently, these DEPs were enriched into metabolism-related pathways, including glycolysis/gluconeogenesis, glycosylcarbon metabolism, pyruvate metabolism, and amino acid biosynthesis, etc. The proteins include ALDOA, LDHA, PGK1, PKM, TPI1 are a key note in the PPI network. HIF-1 pathway is also shown in PPI network.
By using the ClueGO for functional enrichment, the DEPs were mainly related to the pathway of aerobic glycolysis, the release of mitochondrial cytochrome C, and RAC1/PAK1/MMP2 pathway (Figures 3E,F; Supplementary Figure S1). Aerobic glycolysis contains the above metabolic pathways such as glycolysis, gluconeogenesis, pyruvate metabolism, and HIF-1 related pathways (Supplementary Figure S1). The results showed that NaAsO2 treatment caused significant changes in metabolism-related pathways in L-02 cells.
Protein Expression Profiles of DIP + As-Treated Group Compared With As-Treated Group (DIP + As/As Group)
Seventy-one (71) DEPs were identified in the DIP + As/As group. By GO analysis, BP-associated DEPs involve primarily SRP-dependent co-translational protein targeting and membrane, translation, cytoplasmic translation, and mitochondrial gene expression (Figure 4A; Supplementary Table S6). They were widely distributed in cells (Figure 4B,C; Supplementary Table S7) and MF is presented in Supplementary Table S8. Of note, combined GO analysis with UniProt annotation, 21 DEPs were ribosomal proteins (RPs), including DAP3, MRPL11, MRPL39, MRPL43, MRPS28, RPL13, RPL17, RPL19, RPL23A, RPL24, RPL26, RPL27, RPL31, RPL6, RPL7, RPL34, RPL35A, RPS13, RPS15A, RPS19, and RPS5. Among them, 5 DEPs were mitochondrial ribosomal proteins, including DAP3, MRPL11, MRPL39, MRPL43, and MRPS28 (Supplementary Table S7). Based on GO analysis and UniProt annotation, the other 16 were classified as cytoplasmic RPs in the present study. KEGG analysis showed the enrichment of ribosome-related protein translation pathways, nucleotides, and protein metabolism pathways (Figure 4C; Supplementary Table S9), which also are consistent with the GO analysis. In the PPI network analysis, a notable feature was that the DEPs associated with ribosomes were enriched. TCA cycle and amino acid-related biosynthesis pathways were also involved (Figure 4D). Similarly, by using ClueGO for functional enrichment analysis, the DEPs in the DIP + As/As group were highly correlated with the ribosomes (Figures 4E,F; Supplementary Figure S2).
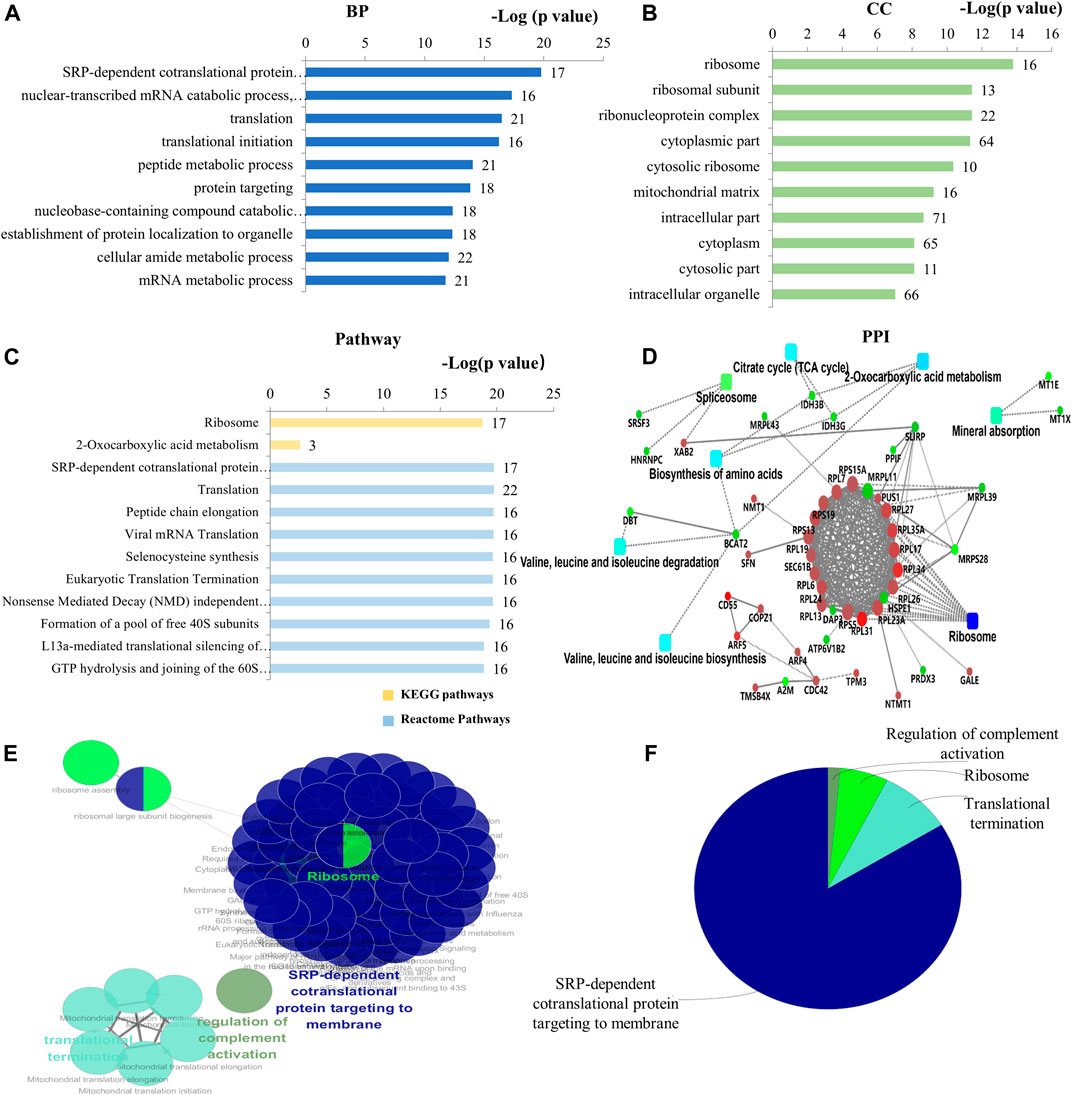
FIGURE 4. Bioinformatics analysis of the deferentially expressed proteins in DIP + As/As group. (A) The top 10 ranking of BP related to the deferentially expressed proteins. (B) The top 10 ranking of CC related to the deferentially expressed proteins. (C) The significantly enriched pathways associated with the deferentially expressed proteins. KEGG pathway and the top 10 ranking of Reactome pathways. (D) PPI network linked to the differentially expressed proteins in DIP + As/As group. Red: the expression of these proteins was up-regulated. Green: the expression of these proteins was down-regulated. (E) Functional interaction network analysis was performed by using ClueGO cytoscape plugin. (F) Overview the specific cluster. (A–C) Number of proteins associated with each category for gene-enrichment analysis is shown on the right of each term bar.
Protein Expression Profiles of DIP + As-Treated Group Compared With Control Group (DIP + As/Ctrl Group) and the Reversed Proteins Between As/Ctrl Group and DIP + As/As Group
Subsequently, we analyzed the protein expression profiles of DIP + As-treated group compared with the control group. Thirteen (13) DEPs were identified in this group. The relatively small number of DEPs suggested that DIP pre-intervention altered the protein expression profile of L-02 cells exposed to As, making it similar to normal growth L-02 cells. In addition, a total of 14 DEPs overlapped between As/Ctrl group and DIP + As/As group. Among these, 9 proteins were up-regulated and 5 proteins were down-regulated in As/Ctrl group, while their expression trend was reversed in DIP + As/As group. They were named as revered proteins (Supplementary Figure S3A).
GO analysis showed that these 14 revered proteins were widely distributed in cells, and KEGG analysis identified that these reversed proteins were not only primarily involved in the regulation of metabolic pathways and other biological processes, including: 2-oxygen carboxylic acid metabolism, amino acid biosynthesis, citric acid cycle (TCA cycle), but also related to the p53 signaling pathway (Supplementary Figure S3B,C; Supplementary Table S10). The PPI network showed that those proteins were correlated with carbon metabolism, amino acid biosynthetic, TCA cycle, 2-oxycarboxylic acid metabolism, cell cycle, and P53 signaling pathways (Supplementary Figure S3D).
Hub Gene Analysis and Cluster Analysis of the Expression of Differentially Expressed Proteins in the Key Pathways
The hub genes in As/Ctrl group and DIP + As/As group were analyzed. As shown in Figure 5A,B, 8 DEPs (PKM, TPI1, ALDOA, PGK1, ALPI, ALPL, MIF, and ANPEP) were identified as hub genes in As/Ctrl group. Twenty-three (23) DEPs were identified as hub genes in DIP + As/As group. Of these, 16 were ribosomal proteins.
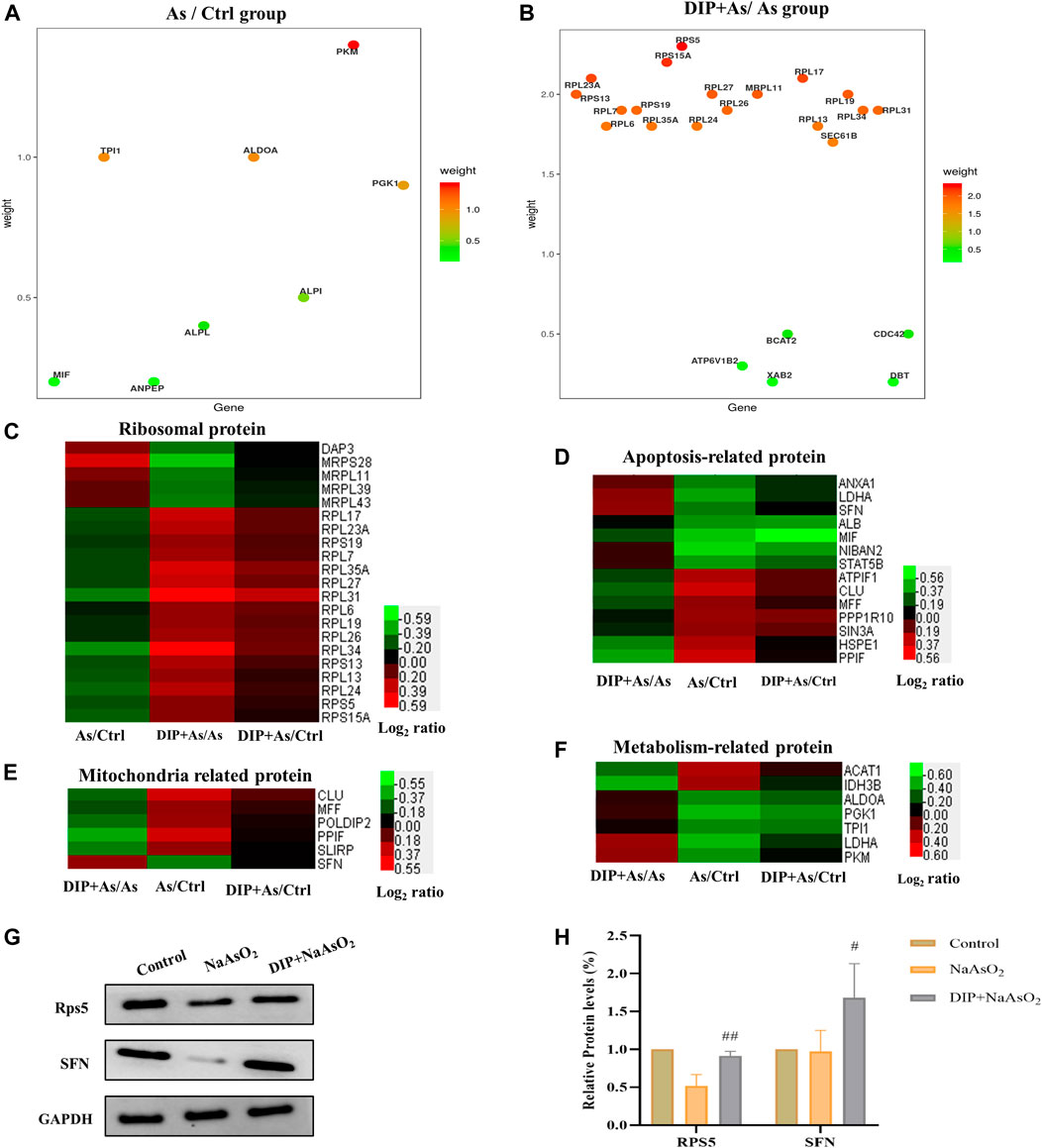
FIGURE 5. Hub gene and cluster analysis and Western blot analysis verification of DEPs in the key pathways. (A) The bubble chart the differentially expressed proteins associated with As/Ctrl group. (B) The bubble chart the differentially expressed proteins associated with DIP + As/As group. (C) Cluster analysis of ribosomal proteins in different groups. (D) Cluster analysis of apoptosis-related proteins in different groups. (E) Cluster analysis of mitochondria related proteins in different groups. (F) Cluster analysis of metabolism-related protein in different groups. (G,H) The differential expression proteins were verified by Western blot analysis.
We also analyzed the expression of DEPs in different groups in the key pathways, i.e., ribosomal protein, apoptosis, mitochondria, and metabolism-related protein (Figures 5C–F). The results showed that DIP pretreatment reversed or restored their expression.
Verification of the Differentially Expressed Proteins by Western Blot Analysis
Based on the above analysis, two hub proteins (RPS5 and SFN), for their largest weight in biological pathways, were selected to be confirmed by Western blot analysis (Figures 5A,B). As shown in Figures 5G,H, consistent with the proteomic results, they were down-regulated in the As/Ctrl group, while DIP pretreatment increased their levels in the DIP + As/As group.
Discussion
Arsenic is one of the most important toxic elements in the natural environment (Dkhil et al., 2020). So far, endemic arsenism is endangering millions of people worldwide. Arsenic toxicity affects almost all organs, of which the liver is one of the main target organs (Jomova et al., 2011; Samelo et al., 2020). Arsenic toxicity can increase the level of ROS in hepatocytes and cause damage to the mitochondrial respiratory chain, destroy the homeostasis of glucose in the liver, and induce oxidative stress (Rezaei et al., 2019), and result in apoptosis towards the cell caused by ROS (Chirumbolo and Bjorklund, 2017). As expected, here, arsenic exposure induced the apoptosis of L-02 cells. After arsenic treatment, the Bax was increased and Bcl-2 was down-regulated, thus facilitating cytochrome c release. Cytochrome c release increases caspase-3 activation, resulting with DNA fragmentation and chromatin concentration, and ultimately inducing apoptosis (Chimenti et al., 2018; Kalpage et al., 2019). However, DIP inhibited the NaAsO2-induced apoptosis in L-02 cells. It antagonized up-regulated Bax expression and down-regulated Bcl-2 expression in L-02 cells exposed to NaAsO2. DIP may serve as a potential scavenger for products of oxidative stress (Deng et al., 2012), thereby reducing ROS levels to ameliorate L-02 cell apoptosis caused by arsenic.
Interestingly, by proteomic analysis, 6 DEPs (CLU, MFF, POLDIP2, PPIF, SFN, and SLIRP) in As/Ctrl group were found to be associated with mitochondrial cytochrome c release, apoptotic mitochondrial changes, and mitochondrion morphogenesis, thereby contributing to sodium arsenite induced apoptosis of L-02 cells. In addition, the DEPs related to metabolic pathways were also identified in this group. The pathways mainly included glycolysis/gluconeogenesis, carbon metabolism, pyruvate metabolism, HIF-1 signaling pathway, and biosynthesis of amino acids. These pathways have been observed in a proteomics study of arsenic-induced liver fibrosis in rats (Wu et al., 2018) or yeast (Guerra-Moreno et al., 2015). We found that most of the related proteins were down-regulated, including ALDOA, LDHA, PGK1, PKM, TPI1. This indicates that after NaAsO2 treatment, the metabolism-related pathway of L-02 cells decreases. It is consistent with previous study that arsenic inhibits ATP production, particularly through glycolysis or pyruvate metabolism (Guerra-Moreno et al., 2015; Wang et al., 2020b). ALDOA and PKM have been observed to be down-regulated in the above arsenic-treated yeast cells study (Wu et al., 2018). Therefore, in the present study, the effect of arsenic tends to reduce the metabolism of L-02 cells. However, low-level arsenic exposure (75 ppb) can induce aerobic glycolysis (Warburg effect), which is a common phenomenon in cultured human primary cells and cell lines (Zhao et al., 2013). Taken together, these results suggest that when arsenic causes apoptosis, it may cause a decrease in cell metabolism, while at lower concentrations it may cause aerobic glycolysis.
Moreover, 14 DEPs in As/Ctrl group were found to be associated with apoptosis, suggesting that they participate in the process. According to Uniprot protein database (https://www.uniprot.org/), in terms of the function of these proteins, they may play a key role in arsenic-induced L-02 apoptosis. For example, SIN3A is a transcriptional repressor; MFF plays a role in the division of mitochondria and peroxisomes; PPIF is involved in the regulation of mitochondrial permeability transition pore (mPTP); PPP1R10 is a scaffold protein, which plays a role in the control of chromatin structure and cell cycle progression. The expression of these 4 proteins increased in the As/Ctrl group, and they may be related to the apoptosis of L-02 cells induced by NaAsO2. On the other hand, the function of STAT5B was signal transduction and transcription activation; NIBAN2 plays a role in the inhibition of apoptosis. Their down-regulation in the As/Ctrl group may also contribute to NaAsO2-induced apoptosis. Interestingly, MFF (Li et al., 2019), PPIF (Folda et al., 2016), and STAT5B (Wetzler et al., 2006) have been reported to be associated with arsenic-induced apoptosis.
Of note, 21 DEPs were identified as ribosomal proteins (RPs). Among them, the expression levels of 16 cytoplasmic RPs showed a downward trend. This is similar to two previous studies on the effect of arsenic on yeast cells (Hosiner et al., 2009; Guerra-Moreno et al., 2015), a large number of ribosomal proteins (74 subunits showed a significant reduction) (Guerra-Moreno et al., 2015) or genes (Hosiner et al., 2009) showed a significant reduction after treatment with arsenic. RPs are essential components of the ribosome that comprise a family of RNA-binding proteins involved in modulating a wide variety of biological processes (Ji et al., 2019). This reduction in ribosome abundance may reflect an adaptive response and is a mechanism that protects cells against its toxicity (Guerra-Moreno et al., 2015). On one hand, ribosome biogenesis is a major consumer of cellular energy and RNA polymerase II activity. The down-regulation of the ribosome itself may lead to energy reserves (Gasch et al., 2000) and the expression of other genes as well (Warner, 1999), such as heat-shock proteins (HSPs) (Hosiner et al., 2009). On the other hand, arsenic induces protein misfolding, by reducing the ribosome level, the production of newly synthesized misfolded proteins can be limited, thus allowing the protein degradation pathway to deal with existing misfolded proteins more effectively (Guerra-Moreno et al., 2015). Interestingly, mutants of the ribosome have been observed to be increased arsenic resistant of yeast cell (Dilda et al., 2008; Hosiner et al., 2009; Guerra-Moreno et al., 2015). Other heavy metals such as mercury, nickel (Hosiner et al., 2009), and cadmium (Guerra-Moreno et al., 2015), as well as multiple stress responses or environmental stress (Gasch et al., 2000), can also affect and down-regulate RPs expression. Therefore, ribosome reduction is not specific to arsenic (Guerra-Moreno et al., 2015) and may be a general feature of the environmental stress (Gasch et al., 2000). In addition to cytoplasmic RPs, mitochondria contain ribosomes that synthesize their own proteins. In yeast cells treated with arsenic, most MPRs showed no change in protein abundance (Guerra-Moreno et al., 2015). However, in this study, five MRPs showed an increased expression trend in As/Ctrl group.
Our results indicate that these changes in RPs may be related to the L-02 cell apoptosis induced by arsenic. In fact, apoptosis is particularly sensitive to nucleolar stress signals, and its primary result is the destruction of ribosome synthesis (Rubbi and Milner, 2003). In ribosomal replication disruption, the free ribosomal proteins interact with the p53 system, leading to cell cycle arrest or apoptosis (Warner and McIntosh, 2009). Increasing evidence shows that inhibiting the expression of RPs, such as knocking down many single RPs, will cause p53 accumulation, thereby leading to cell apoptosis (Uechi et al., 2006; Danilova et al., 2008; Daftuar et al., 2013). Interestingly, arsenic has been shown to cause cell death via a p53-dependent mechanism (Yu et al., 2008). Among these cytoplasmic RPs, 5 RPs have been reported to be associated with apoptosis, including RPL23 (Dai et al., 2004; Jin et al., 2004), RPL34 (Ji et al., 2019), Ll3a (Chen and Ioannou, 1999), L7 (Chen and Ioannou, 1999), and L35a (Lopez et al., 2002). Inhibition of their expression, such as through siRNA-mediated silencing, leads to cell apoptosis or inhibits cell proliferation (Chen and Ioannou, 1999; Lopez et al., 2002; Dai et al., 2004; Jin et al., 2004; Ji et al., 2019). RNAi-mediating silencing of RPS5 gene expression also resulted in the inability of MEL cells to differentiate (Vizirianakis et al., 2015).
In the view of MRPs, their abnormal expression can cause mitochondrial metabolism disorder and cell dysfunction (Huang et al., 2020). Some MRPs such as MRPS29 (DAP3), MRPL41, MRPS30, and MRPL64 have been identified to be associated with apoptosis by p53 pathway (Yoo et al., 2005; Yan et al., 2017; Huang et al., 2020). In this study, three MRPs (i.e., DAP3, MRPL39, and MRPS28) showed an up-regulated trend in As/Ctrl group. They may be related to L02 cell apoptosis induced by NaAsO2. DAP3 (death-associated protein 3), also known as MRPS29; its high expression can promote apoptosis (Miyazaki et al., 2004). Another protein MRPL39 was reported to serve as a tumor suppressor (Yu et al., 2018). Likewise, the inhibition of MRPS28 is related to the treatment of glioblastoma with Benzyl isothiocyanate (BITC) (Tang et al., 2016).
Indeed, the relationship between ribosomes and cell proliferation and apoptosis has been well reviewed (Warner and McIntosh, 2009; Kim et al., 2017; Huang et al., 2020). However, most of the studies on the effects of arsenic on ribosomes involve yeast and tumor cells (Chen and Ioannou, 1999; Gasch et al., 2000; Haugen et al., 2004; Hosiner et al., 2009; Guerra-Moreno et al., 2015; Kim et al., 2017). To the best of our knowledge, the present study is the first to report that arsenic induces a broadly decrease in cytoplasmic RPs, and an increase in mitochondrial RPs in liver cells, resulting in cell apoptosis. Our finding supports this opinion that a number of RPs have secondary roles regardless of their presence in protein biosynthesis, regulation of cell proliferation, or in some cases acting as inducers of cell death (Chen and Ioannou, 1999). Interestingly, the expression of these RPs was reversed in the DIP + As/Ctrl group, suggesting that they may be responsible for DIP attenuating arsenic-induced apoptosis of L-02 cells.
Furthermore, we noted that several proteins may play important roles in arsenic-induced apoptosis and/or DIP resistance to arsenic-induced apoptosis, including ALPI, ALPL CDC42, EIF5A, HSPE1, and SFN. Arsenic sensitivity to enhanced alkaline phosphatase (ALP) activity has been reported (Herrera et al., 2013), where alkaline phosphatase ALPI and ALPL have been shown to be up-regulated in treated cells, whereas DIP pretreatment has reversed its expression. CDC42 is a small GTPase of Rho family, involved in regulation of various functions including positive regulation of cell growth. Here, it was up-regulated in DIP + As/Ctrl group and may be linked to the intervention of DIP on arsenic-induced apoptosis. Two translation initiation factors, eIF2E and eIF4E, have been reported to be related to arsenic-induced cytotoxicity and cell death (Othumpangat et al., 2005; Guerra-Moreno et al., 2015). Here, down-regulation of EIF5A in As/Ctrl may contribute to the NaAsO2-induced L-02 cell apoptosis. Arsenic can also cause protein misfolding and induce an increase in HSP protein expression (Tam and Wang, 2020). In this study, the expression of heat shock protein HSPE1 increased in As/Ctrl group. It may participate in endoplasmic reticulum stress and unfolded protein response, and remove misfolded proteins induced by arsenic. SFN (14-3-3 protein Sigma) is one of the isoforms of the 14-3-3 family. This family of proteins regulates a variety of cell functions (Lee et al., 2014). Overexpression of SFN in multiple myeloma cells attenuated arsenic trioxide-induced cell death (Ge et al., 2009). In this study, it was down-regulated in the As/Ctrl group and up-regulated in the DIP + As/Ctrl group, suggesting that it may be involved in NaAsO2-induced apoptosis and DIP attenuated apoptosis.
Conclusion
In this study, apoptosis can be induced in human normal liver cells L-02 cells using NaAsO2. When hepatic cells were pretreated, the apoptosis was reduced. DIP inhibits cell apoptosis and was associated with an increase in Bcl-2 anti-apoptotic protein and a decrease in Bax pro-apoptotic protein. This may be due to the DIP antioxidant. Proteomic analysis showed that metabolism, apoptosis, and mitochondria-related proteins were associated with arsenic induced apoptosis of L-02 cells. Arsenic concentration induced apoptosis inhibited aerobic glycolysis of L-02 cells. DIP pretreatment reversed or restored the expression of these proteins, suggesting that they were linked to the prevention of DIP apoptosis. Importantly, this is the first study to observe that extensive variations in RPs were associated with arsenic-induced apoptosis in normal human cells. Cytoplasmic ribosomes were down-regulated and mitochondrial ribosomes were up-regulated. The expression of these proteins was reversed by DIP pretreatment. This is also the first study to relate the influence of DIP on apoptosis and its ability to control RP expression. The pathways by which arsenic induces L-02 cell apoptosis and DIP attenuate arsenic-induced apoptosis are summarized in Figure 6. Moreover, there are limitations in this study. It was carried out on a single cell line and needs to be further verified in more cell lines or biological tissues. The mechanism of arsenic-induced apoptosis and DIP’s inhibition of apoptosis disclosed in this study need to be further clarified. We will also explore the quantity (minimum inhibitory concentration) required for DIP to have a protective effect compared to other biological products.
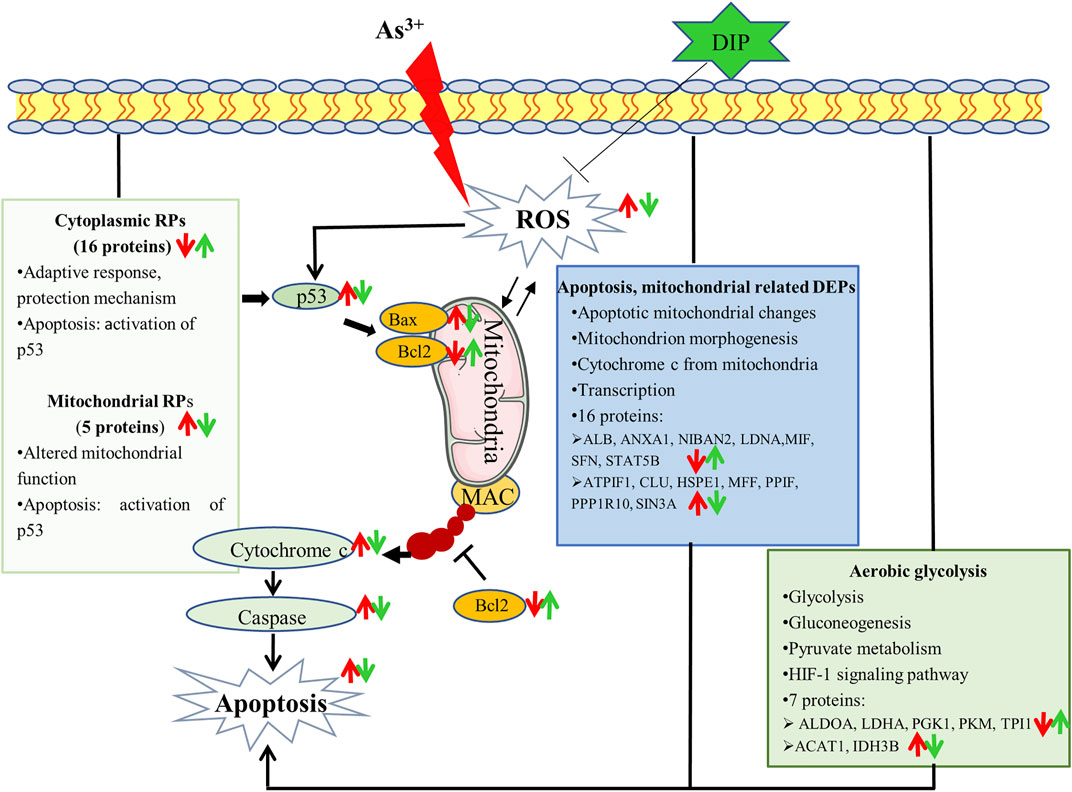
FIGURE 6. Potential mechanism that sodium arsenite-induced L-02 cell apoptosis and DIP attenuates sodium arsenite-induced apoptosis.: ↑up-regulation.: ↓down-regulation. Red is related to As3+, green is related to DIP.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author Contributions
All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
Funding
This research was supported by the National Natural Science Foundation of China (approval numbers: 81660835, 81860560), the Foundation of Guizhou Educational Committee (No. KY (2021) 008), the National Natural Science Foundation of China (U1812403-6-2-4), and Shenzhen-Hong Kong Institute of Brain Science-Shenzhen Fundamental Research Institutions (2021SHIBS0003).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We are grateful to the instrument analysis center of Shenzhen University and the following research institutions for their platform support for this research: Guizhou Medical University School of Public Health; Guizhou Medical University Environmental Pollution Monitoring and Control Ministry of Education Key Laboratory; State Key Laboratory of Functions and Applications of Medicinal Plants, Guizhou Medical University; Guizhou Provincial Food Nutrition and Health Engineering Research Center.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphar.2021.749035/full#supplementary-material
Supplementary Figure S1 | Specific cluster of ClueGO analysis for the As/Ctrl group.
Supplementary Figure S2 | Specific cluster of ClueGO analysis for DIP+As/As group.
Supplementary Figure S3 | Bioinformatics analysis of the reversed proteins between As/Ctrl, and DIP+As/As group. (A) Cluster analysis of the reversed proteins by using Cluster 3.0. (B) The GO annotations of the reversed proteins. (C) The KEGG pathways associated with the reversed proteins. (D) PPI network analysis of the reversed proteins.
References
Adil, M., Kandhare, A. D., Ghosh, P., and Bodhankar, S. L. (2016). Sodium Arsenite-Induced Myocardial Bruise in Rats: Ameliorative Effect of Naringin via TGF-β/Smad and Nrf/HO Pathways. Chem. Biol. Interact 253, 66–77. doi:10.1016/j.cbi.2016.05.015
Adil, M., Kandhare, A. D., Visnagri, A., and Bodhankar, S. L. (2015). Naringin Ameliorates Sodium Arsenite-Induced Renal and Hepatic Toxicity in Rats: Decisive Role of KIM-1, Caspase-3, TGF-β, and TNF-α. Ren. Fail. 37 (8), 1396–1407. doi:10.3109/0886022X.2015.1074462
Bashir, S., Sharma, Y., Irshad, M., Nag, T. C., Tiwari, M., Kabra, M., et al. (2006). Arsenic Induced Apoptosis in Rat Liver Following Repeated 60 Days Exposure. Toxicology 217 (1), 63–70. doi:10.1016/j.tox.2005.08.023
Bhattacharya, S., Das, S. K., and Haldar, P. K. (2014). Arsenic Induced Myocardial Toxicity in Rats: Alleviative Effect of Trichosanthes Dioica Fruit. J. Diet. Suppl. 11 (3), 248–261. doi:10.3109/19390211.2014.937044
Bindea, G., Mlecnik, B., Hackl, H., Charoentong, P., Tosolini, M., Kirilovsky, A., et al. (2009). ClueGO: a Cytoscape Plug-In to Decipher Functionally Grouped Gene Ontology and Pathway Annotation Networks. Bioinformatics 25 (8), 1091–1093. doi:10.1093/bioinformatics/btp101
Chen, F. W., and Ioannou, Y. A. (1999). Ribosomal Proteins in Cell Proliferation and Apoptosis. Int. Rev. Immunol. 18 (5-6), 429–448. doi:10.3109/08830189909088492
Chen, H., Hao, Y., Wang, L., Jia, D., Ruan, Y., and Gu, J. (2012). Sodium Arsenite Down-Regulates the Expression of X-Linked Inhibitor of Apoptosis Protein via Translational and post-translational Mechanisms in Hepatocellular Carcinoma. Biochem. Biophys. Res. Commun. 422 (4), 721–726. doi:10.1016/j.bbrc.2012.05.066
Chimenti, M. S., Sunzini, F., Fiorucci, L., Botti, E., Fonti, G. L., Conigliaro, P., et al. (2018). Potential Role of Cytochrome C and Tryptase in Psoriasis and Psoriatic Arthritis Pathogenesis: Focus on Resistance to Apoptosis and Oxidative Stress. Front. Immunol. 9, 2363. doi:10.3389/fimmu.2018.02363
Chirumbolo, S., and Bjørklund, G. (2017). PERM Hypothesis: The Fundamental Machinery Able to Elucidate the Role of Xenobiotics and Hormesis in Cell Survival and Homeostasis. Int. J. Mol. Sci. 18 (1). doi:10.3390/ijms18010165
Daftuar, L., Zhu, Y., Jacq, X., and Prives, C. (2013). Ribosomal Proteins RPL37, RPS15 and RPS20 Regulate the Mdm2-P53-MdmX Network. PLoS One 8 (7), e68667. doi:10.1371/journal.pone.0068667
Dai, M. S., Zeng, S. X., Jin, Y., Sun, X. X., David, L., and Lu, H. (2004). Ribosomal Protein L23 Activates P53 by Inhibiting MDM2 Function in Response to Ribosomal Perturbation but Not to Translation Inhibition. Mol. Cel Biol 24 (17), 7654–7668. doi:10.1128/MCB.24.17.7654-7668.2004
Danilova, N., Sakamoto, K. M., and Lin, S. (2008). Ribosomal Protein S19 Deficiency in Zebrafish Leads to Developmental Abnormalities and Defective Erythropoiesis through Activation of P53 Protein Family. Blood 112 (13), 5228–5237. doi:10.1182/blood-2008-01-132290
Dash, M., Maity, M., Dey, A., Perveen, H., Khatun, S., Jana, L., et al. (2018). The Consequence of NAC on Sodium Arsenite-Induced Uterine Oxidative Stress. Toxicol. Rep. 5, 278–287. doi:10.1016/j.toxrep.2018.02.003
Deng, C., Hu, Z., Fu, H., Hu, M., Xu, X., and Chen, J. (2012). Chemical Analysis and Antioxidant Activity In Vitro of a β-D-glucan Isolated from Dictyophora Indusiata. Int. J. Biol. Macromol 51 (1-2), 70–75. doi:10.1016/j.ijbiomac.2012.05.001
Dilda, P. J., Perrone, G. G., Philp, A., Lock, R. B., Dawes, I. W., and Hogg, P. J. (2008). Insight into the Selectivity of Arsenic Trioxide for Acute Promyelocytic Leukemia Cells by Characterizing Saccharomyces cerevisiae Deletion Strains that Are Sensitive or Resistant to the Metalloid. Int. J. Biochem. Cel Biol 40 (5), 1016–1029. doi:10.1016/j.biocel.2007.11.002
Dkhil, M. A., Abdel Moneim, A. E., Bauomy, A. A., Khalil, M., Al-Shaebi, E. M., and Al-Quraishy, S. (2020). Chlorogenic Acid Prevents Hepatotoxicity in Arsenic-Treated Mice: Role of Oxidative Stress and Apoptosis. Mol. Biol. Rep. 47 (2), 1161–1171. doi:10.1007/s11033-019-05217-4
Duan, T., Hu, T., Wu, C., Yeh, Y. T., Lu, J., Zhang, Q., et al. (2020). PINK1/Parkin-mediated Mitophagy Is Involved in NaAsO2-Induced Apoptosis of Human Hepatic Cells through Activation of ERK Signaling. Toxicol. Vitro 66, 104857. doi:10.1016/j.tiv.2020.104857
Folda, A., Citta, A., Scalcon, V., Calì, T., Zonta, F., Scutari, G., et al. (2016). Mitochondrial Thioredoxin System as a Modulator of Cyclophilin D Redox State. Sci. Rep. 6, 23071. doi:10.1038/srep23071
Gasch, A. P., Spellman, P. T., Kao, C. M., Carmel-Harel, O., Carmel-Harel, M. B., Storz, G., et al. (2000). Genomic Expression Programs in the Response of Yeast Cells to Environmental Changes. MBoC 11 (12), 4241–4257. doi:10.1091/mbc.11.12.4241
Ge, F., Lu, X. P., Zeng, H. L., He, Q. Y., Xiong, S., Jin, L., et al. (2009). Proteomic and Functional Analyses Reveal a Dual Molecular Mechanism Underlying Arsenic-Induced Apoptosis in Human Multiple Myeloma Cells. J. Proteome Res. 8 (6), 3006–3019. doi:10.1021/pr9001004
Goudarzi, M., Fatemi, I., Siahpoosh, A., Sezavar, S. H., Mansouri, E., and Mehrzadi, S. (2018). Protective Effect of Ellagic Acid against Sodium Arsenite-Induced Cardio- and Hematotoxicity in Rats. Cardiovasc. Toxicol. 18 (4), 337–345. doi:10.1007/s12012-018-9446-2
Guerra-Moreno, A., Isasa, M., Bhanu, M. K., Waterman, D. P., Eapen, V. V., Gygi, S. P., et al. (2015). Proteomic Analysis Identifies Ribosome Reduction as an Effective Proteotoxic Stress Response. J. Biol. Chem. 290 (50), 29695–29706. doi:10.1074/jbc.M115.684969
Habtemariam, S. (2019). The Chemistry, Pharmacology and Therapeutic Potential of the Edible Mushroom Dictyophora Indusiata (Vent Ex. Pers.) Fischer (Synn. Phallus Indusiatus). Biomedicines 7 (4). doi:10.3390/biomedicines7040098
Han, S., Ma, C., Hu, M., Wang, Y., Ma, F., Tao, N., et al. (2017). A Polysaccharide from Dictyophora Indusiata Inhibits the Immunosuppressive Function of Cancer-Associated Fibroblasts. Cell Biochem Funct 35 (7), 414–419. doi:10.1002/cbf.3290
Haugen, A. C., Kelley, R., Collins, J. B., Tucker, C. J., Deng, C., Afshari, C. A., et al. (2004). Integrating Phenotypic and Expression Profiles to Map Arsenic-Response Networks. Genome Biol. 5 (12), R95. doi:10.1186/gb-2004-5-12-r95
Herrera, A., Pineda, J., and Antonio, M. T. (2013). Toxic Effects of Perinatal Arsenic Exposure on the Brain of Developing Rats and the Beneficial Role of Natural Antioxidants. Environ. Toxicol. Pharmacol. 36 (1), 73–79. doi:10.1016/j.etap.2013.03.018
Hosiner, D., Lempiäinen, H., Reiter, W., Urban, J., Loewith, R., Ammerer, G., et al. (2009). Arsenic Toxicity to Saccharomyces cerevisiae Is a Consequence of Inhibition of the TORC1 Kinase Combined with a Chronic Stress Response. Mol. Biol. Cel 20 (3), 1048–1057. doi:10.1091/mbc.E08-04-0438
Hu, T., Zhang, K., Pan, D., Pan, X., Yang, H., Xiao, J., et al. (2020). Inhibition Effect of Dictyophora Polysaccharides on Human Hepatocellular Carcinoma Cell Line HCC-LM3. Med. Sci. Monit. 26, e918870. doi:10.12659/MSM.918870
Huang, G., Li, H., and Zhang, H. (2020). Abnormal Expression of Mitochondrial Ribosomal Proteins and Their Encoding Genes with Cell Apoptosis and Diseases. Int. J. Mol. Sci. 21 (22). doi:10.3390/ijms21228879
Iqbal, J., Zhang, K., Jin, N., Zhao, Y., Liu, Q., Ni, J., et al. (2018). Effect of Sodium Selenate on Hippocampal Proteome of 3×Tg-AD Mice-Exploring the Antioxidant Dogma of Selenium against Alzheimer's Disease. ACS Chem. Neurosci. 9 (7), 1637–1651. doi:10.1021/acschemneuro.8b00034
Ji, P., Wang, L., Liu, J., Mao, P., Li, R., Jiang, H., et al. (2019). Knockdown of RPL34 Inhibits the Proliferation and Migration of Glioma Cells through the Inactivation of JAK/STAT3 Signaling Pathway. J. Cel Biochem 120 (3), 3259–3267. doi:10.1002/jcb.27592
Jin, A., Itahana, K., O'Keefe, K., and Zhang, Y. (2004). Inhibition of HDM2 and Activation of P53 by Ribosomal Protein L23. Mol. Cel Biol 24 (17), 7669–7680. doi:10.1128/MCB.24.17.7669-7680.2004
Jomova, K., Jenisova, Z., Feszterova, M., Baros, S., Liska, J., Hudecova, D., et al. (2011). Arsenic: Toxicity, Oxidative Stress and Human Disease. J. Appl. Toxicol. 31 (2), 95–107. doi:10.1002/jat.1649
Kalpage, H. A., Bazylianska, V., Recanati, M. A., Fite, A., Liu, J., Wan, J., et al. (2019). Tissue-specific Regulation of Cytochrome C by post-translational Modifications: Respiration, the Mitochondrial Membrane Potential, ROS, and Apoptosis. FASEB J. 33 (2), 1540–1553. doi:10.1096/fj.201801417R
Kanwal, S., Joseph, T. P., Owusu, L., Xiaomeng, R., Meiqi, L., and Yi, X. (2018). A Polysaccharide Isolated from Dictyophora Indusiata Promotes Recovery from Antibiotic-Driven Intestinal Dysbiosis and Improves Gut Epithelial Barrier Function in a Mouse Model. Nutrients 10 (8). doi:10.3390/nu10081003
Kim, H. J., Maiti, P., and Barrientos, A. (2017). Mitochondrial Ribosomes in Cancer. Semin. Cancer Biol. 47, 67–81. doi:10.1016/j.semcancer.2017.04.004
Lee, Y. K., Hur, W., Lee, S. W., Hong, S. W., Kim, S. W., Choi, J. E., et al. (2014). Knockdown of 14-3-3ζ Enhances Radiosensitivity and Radio-Induced Apoptosis in CD133(+) Liver Cancer Stem Cells. Exp. Mol. Med. 46, e77. doi:10.1038/emm.2013.151
Li, S., Zhao, H., Wang, Y., Shao, Y., Liu, J., and Xing, M. (2019). Arsenic-induced Cardiotoxicity Correlates with Mitochondrial Damage and Trace Elements Imbalance in Broiler Chickens. Poult. Sci. 98 (2), 734–744. doi:10.3382/ps/pey469
Liao, W., Yu, Z., Lin, Z., Lei, Z., Ning, Z., Regenstein, J. M., et al. (2015). Biofunctionalization of Selenium Nanoparticle with Dictyophora Indusiata Polysaccharide and its Antiproliferative Activity through Death-Receptor and Mitochondria-Mediated Apoptotic Pathways. Sci. Rep. 5, 18629. doi:10.1038/srep18629
Lin, A., Zhang, X., Zhu, Y. G., and Zhao, F. J. (2008). Arsenate-induced Toxicity: Effects on Antioxidative Enzymes and DNA Damage in Vicia faba. Environ. Toxicol. Chem. 27 (2), 413–419. doi:10.1897/07-266R.1
Lopez, C. D., Martinovsky, G., and Naumovski, L. (2002). Inhibition of Cell Death by Ribosomal Protein L35a. Cancer Lett. 180 (2), 195–202. doi:10.1016/s0304-3835(02)00024-1
Lv, Y., Hu, Q., Shi, M., Wang, W., Zheng, Y., Yang, Z., et al. (2020). The Role of PSMB5 in Sodium Arsenite-Induced Oxidative Stress in L-02 Cells. Cell Stress Chaperones 25 (3), 533–540. doi:10.1007/s12192-020-01104-1
Miyazaki, T., Shen, M., Fujikura, D., Tosa, N., Kim, H. R., Kon, S., et al. (2004). Functional Role of Death-Associated Protein 3 (DAP3) in Anoikis. J. Biol. Chem. 279 (43), 44667–44672. doi:10.1074/jbc.M408101200
Ola-Davies, O. E., and Akinrinde, A. S. (2016). Acute Sodium Arsenite-Induced Hematological and Biochemical Changes in Wistar Rats: Protective Effects of Ethanol Extract of Ageratum Conyzoides. Pharmacognosy Res. 8 (Suppl. 1), S26–S30. doi:10.4103/0974-8490.178645
Othumpangat, S., Kashon, M., and Joseph, P. (2005). Sodium Arsenite-Induced Inhibition of Eukaryotic Translation Initiation Factor 4E (eIF4E) Results in Cytotoxicity and Cell Death. Mol. Cel Biochem 279 (1-2), 123–131. doi:10.1007/s11010-005-8284-2
Perker, M. C., Orta Yilmaz, B., Yildizbayrak, N., Aydin, Y., and Erkan, M. (2019). Protective Effects of Curcumin on Biochemical and Molecular Changes in Sodium Arsenite-Induced Oxidative Damage in Embryonic Fibroblast Cells. J. Biochem. Mol. Toxicol. 33 (7), e22320. doi:10.1002/jbt.22320
Rezaei, M., Keshtzar, E., Khodayar, M. J., and Javadipour, M. (2019). SirT3 Regulates Diabetogenic Effects Caused by Arsenic: An Implication for Mitochondrial Complex II Modification. Toxicol. Lett. 301, 24–33. doi:10.1016/j.toxlet.2018.10.025
Saha, S., Rashid, K., Sadhukhan, P., Agarwal, N., and Sil, P. C. (2016). Attenuative Role of Mangiferin in Oxidative Stress-Mediated Liver Dysfunction in Arsenic-Intoxicated Murines. Biofactors 42 (5), 515–532. doi:10.1002/biof.1276
Samelo, R. R., da Cunha de Medeiros, P., de Carvalho Cavalcante, D. N., Aranha, M. L. G., Duarte, F. A., de Castro, Í. B., et al. (2020). Low Concentrations of Sodium Arsenite Induce Hepatotoxicity in Prepubertal Male Rats. Environ. Toxicol. 35 (5), 553–560. doi:10.1002/tox.22890
Shen, L., Yang, A., Chen, X., Xiao, S., Liu, X., Lin, J., et al. (2021). Proteomic Profiling of Cerebrum Mitochondria, Myelin Sheath, and Synaptosome Revealed Mitochondrial Damage and Synaptic Impairments in Association with 3 × Tg‐AD Mice Model. Cell Mol Neurobiol. doi:10.1007/s10571-021-01052-z
Sun, H., Yang, Y., Shao, H., Sun, W., Gu, M., Wang, H., et al. (2017). Sodium Arsenite-Induced Learning and Memory Impairment Is Associated with Endoplasmic Reticulum Stress-Mediated Apoptosis in Rat Hippocampus. Front. Mol. Neurosci. 10, 286. doi:10.3389/fnmol.2017.00286
Tam, L. M., and Wang, Y. (2020). Arsenic Exposure and Compromised Protein Quality Control. Chem. Res. Toxicol. 33 (7), 1594–1604. doi:10.1021/acs.chemrestox.0c00107
Tang, N. Y., Chueh, F. S., Yu, C. C., Liao, C. L., Lin, J. J., Hsia, T. C., et al. (2016). Benzyl Isothiocyanate Alters the Gene Expression with Cell Cycle Regulation and Cell Death in Human Brain Glioblastoma GBM 8401 Cells. Oncol. Rep. 35 (4), 2089–2096. doi:10.3892/or.2016.4577
Uechi, T., Nakajima, Y., Nakao, A., Torihara, H., Chakraborty, A., Inoue, K., et al. (2006). Ribosomal Protein Gene Knockdown Causes Developmental Defects in Zebrafish. PLoS One 1, e37. doi:10.1371/journal.pone.0000037
Vizirianakis, I. S., Papachristou, E. T., Andreadis, P., Zopounidou, E., Matragkou, C. N., and Tsiftsoglou, A. S. (2015). Genetic Manipulation of RPS5 Gene Expression Modulates the Initiation of Commitment of MEL Cells to Erythroid Maturation: Implications in Understanding Ribosomopathies. Int. J. Oncol. 47 (1), 303–314. doi:10.3892/ijo.2015.3017
Wang, W., Liu, H., Zhang, Y., Feng, Y., Yuan, F., Song, X., et al. (2019a). Antihyperlipidemic and Hepatoprotective Properties of Alkali- and Enzyme-Extractable Polysaccharides by Dictyophora Indusiata. Sci. Rep. 9 (1), 14266. doi:10.1038/s41598-019-50717-9
Wang, W., Song, X., Zhang, J., Li, H., Liu, M., Gao, Z., et al. (2019b). Antioxidation, Hepatic- and Renal-protection of Water-Extractable Polysaccharides by Dictyophora Indusiata on Obese Mice. Int. J. Biol. Macromol 134, 290–301. doi:10.1016/j.ijbiomac.2019.05.028
Wang, W., Wang, Q., Zou, Z., Zheng, F., and Zhang, A. (2020a). Human Arsenic Exposure and Lung Function Impairment in Coal-Burning Areas in Guizhou, China. Ecotoxicol Environ. Saf. 190, 110174. doi:10.1016/j.ecoenv.2020.110174
Wang, W., Zheng, F., Lin, C., and Zhang, A. (2020b). Changes in Energy Metabolism and Macrophage Polarization: Potential Mechanisms of Arsenic-Induced Lung Injury. Ecotoxicol Environ. Saf. 204, 110948. doi:10.1016/j.ecoenv.2020.110948
Wang, Y., Ji, X., Yan, M., Chen, X., Kang, M., Teng, L., et al. (2019c). Protective Effect and Mechanism of Polysaccharide from Dictyophora Indusiata on Dextran Sodium Sulfate-Induced Colitis in C57BL/6 Mice. Int. J. Biol. Macromol 140, 973–984. doi:10.1016/j.ijbiomac.2019.08.198
Warner, J. R., and McIntosh, K. B. (2009). How Common Are Extraribosomal Functions of Ribosomal Proteins?. Mol. Cel 34 (1), 3–11. doi:10.1016/j.molcel.2009.03.006
Warner, J. R. (1999). The Economics of Ribosome Biosynthesis in Yeast. Trends Biochem. Sci. 24 (11), 437–440. doi:10.1016/s0968-0004(99)01460-7
Wei, Y., Zeng, B., Zhang, H., Chen, C., Wu, Y., Wang, N., et al. (2018). Comparative Proteomic Analysis of Fluoride Treated Rat Bone Provides New Insights into the Molecular Mechanisms of Fluoride Toxicity. Toxicol. Lett. 291, 39–50. doi:10.1016/j.toxlet.2018.04.006
Wetzler, M., Brady, M. T., Tracy, E., Li, Z. R., Donohue, K. A., O'Loughlin, K. L., et al. (2006). Arsenic Trioxide Affects Signal Transducer and Activator of Transcription Proteins through Alteration of Protein Tyrosine Kinase Phosphorylation. Clin. Cancer Res. 12 (22), 6817–6825. doi:10.1158/1078-0432.CCR-06-1354
Wu, S., Li, J., and Jin, X. (2018). iTRAQ-Based Quantitative Proteomic Analysis Reveals Important Metabolic Pathways for Arsenic-Induced Liver Fibrosis in Rats. Sci. Rep. 8 (1), 3267. doi:10.1038/s41598-018-21580-x
Yan, H. X., Zhang, Y. J., Zhang, Y., Ren, X., Shen, Y. F., Cheng, M. B., et al. (2017). CRIF1 Enhances P53 Activity via the Chromatin Remodeler SNF5 in the HCT116 colon Cancer Cell Lines. Biochim. Biophys. Acta Gene Regul. Mech. 1860 (4), 516–522. doi:10.1016/j.bbagrm.2017.02.006
Yoo, Y. A., Kim, M. J., Park, J. K., Chung, Y. M., Lee, J. H., Chi, S. G., et al. (2005). Mitochondrial Ribosomal Protein L41 Suppresses Cell Growth in Association with P53 and p27Kip1. Mol. Cel Biol 25 (15), 6603–6616. doi:10.1128/MCB.25.15.6603-6616.2005
Yu, M. J., Zhao, N., Shen, H., and Wang, H. (2018). Long Noncoding RNA MRPL39 Inhibits Gastric Cancer Proliferation and Progression by Directly Targeting miR-130. Genet. Test. Mol. Biomarkers 22 (11), 656–663. doi:10.1089/gtmb.2018.0151
Yu, W. X., Lin, C. Q., Zhao, Q., Lin, X. J., and Dong, X. L. (2017). Neuroprotection against Hydrogen Peroxide-Induced Toxicity by Dictyophora Echinovolvata Polysaccharide via Inhibiting the Mitochondria-dependent Apoptotic Pathway. Biomed. Pharmacother. 88, 569–573. doi:10.1016/j.biopha.2017.01.103
Yu, X., Robinson, J. F., Gribble, E., Hong, S. W., Sidhu, J. S., and Faustman, E. M. (2008). Gene Expression Profiling Analysis Reveals Arsenic-Induced Cell Cycle Arrest and Apoptosis in P53-Proficient and P53-Deficient Cells through Differential Gene Pathways. Toxicol. Appl. Pharmacol. 233 (3), 389–403. doi:10.1016/j.taap.2008.09.016
Zhang, J., Shi, R., Li, H., Xiang, Y., Xiao, L., Hu, M., et al. (2016). Antioxidant and Neuroprotective Effects of Dictyophora Indusiata Polysaccharide in Caenorhabditis elegans. J. Ethnopharmacol 192, 413–422. doi:10.1016/j.jep.2016.09.031
Keywords: dictyophora polysaccharides, NaAsO2, apoptosis, hepatotoxicity, proteomics
Citation: Hu T, Shen L, Huang Q, Wu C, Zhang H, Zeng Q, Wang G, Wei S, Zhang S, Zhang J, Khan NU, Shen X and Luo P (2021) Protective Effect of Dictyophora Polysaccharides on Sodium Arsenite-Induced Hepatotoxicity: A Proteomics Study. Front. Pharmacol. 12:749035. doi: 10.3389/fphar.2021.749035
Received: 28 July 2021; Accepted: 07 October 2021;
Published: 26 November 2021.
Edited by:
Abdel Halim Harrath, King Saud University, Saudi ArabiaReviewed by:
Mohamed M. Abdel-Daim, Suez Canal University, EgyptImene Hadda Ouzari, Physiques et Naturelles de Tunis, Tunisia
Copyright © 2021 Hu, Shen, Huang, Wu, Zhang, Zeng, Wang, Wei, Zhang, Zhang, Khan, Shen and Luo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiangchun Shen, shenxiangchun@126.com; Peng Luo, luopeng@gmc.edu.cn
†These authors have contributed equally to this work
 Ting Hu
Ting Hu Liming Shen
Liming Shen Qun Huang2,3
Qun Huang2,3 Xiangchun Shen
Xiangchun Shen Peng Luo
Peng Luo