- 1The key Laboratory of Environmental Pollution Monitoring and Disease Control, Ministry of Education of Guizhou and Key Laboratory of Medical Microbiology and Parasitology, School of Basic Medical Sciences, Guizhou Medical University, Guiyang, China
- 2Center of Expertise in Mycology, Radboud University Medical Center/Canisius Wilhelmina Hospital, Nijmegen, Netherlands
- 3Department of Medical Microbiology, The People’s Hospital of Suzhou New District, Suzhou, Jiangsu, China
- 4Natural and Medical Sciences Research Center, University of Nizwa, Nizwa, Oman
- 5Department of Medical Microbiology and Infectious Diseases, Canisius Wilhelmina Hospital, Nijmegen, Netherlands
- 6Bioprocess Engineering and Biotechnology Graduate Program, Federal University of Paraná, Curitiba, Brazil
- 7Department of Microbiology, Muzaffarnagar Medical College, Muzaffarnagar, India
A resistant and hypervirulent dermatophyte from India has been described as a taxonomic novelty, Trichophyton indotineae, a species of the Trichophyton mentagrophytes complex. Rapid detection and correct identification of closely similar dermatophytes with different predilections are essential for efficient clinical management. We evaluated the efficacy of rapid diagnostic methods clinical and environmental strains in the T. mentagrophytes complex. The methods included Real-time-PCR, DermaGenius, LAMP, and MALDI-ToF MS, using rDNA ITS sequences as taxonomic standard. The results show that only MALDI-ToF MS can distinguish 96.97% T. indotineae from other closely related species. The complex comprises numerous clones which may differ in anonymous markers but with similar evolutionary behavior. Therefore, we recommend to distinguish species only when they show an appreciable degree of adaptation and thus are clinically significant. The distinction of remaining clonal diversity is an epidemiological query and can be solved by haplotype numbering.
Introduction
Dermatophytoses are among the most common fungal infections, affecting 20–25% of the world’s population (Havlickova et al., 2008; Ameen, 2010). The species spectrum of dermatophyte infections has changed dramatically over time with socioeconomic developments (Zhan et al., 2018). Most pathogenic agents of humans originated from domesticated animals such as cattle, horse or camels (Deng et al., 2008; Morrell and Stratman, 2011; Hameed et al., 2017). Infections by zoophilic dermatophytes cause highly inflammatory lesions due to immunopathogenesis (Drouot et al., 2009; Celestrino et al., 2021; Sardana et al., 2021). The spectrum of infections of urban populations has changed in favor of species with human-to-human transmission and milder clinical pictures, in addition to those by pet-associated species causing self-limiting outbreaks. It has previously been suggested that long-term alterations of human-animal relationships such as domestication leads to novel lines of evolution in dermatophytes (Tang et al., 2021). This is inherent to dermatophyte natural life cycles, which comprise elaborate sexual reproduction in the environment and asymptomatic carriage of spores in mammalian fur. Domestication interrupts the sexual part of this life cycle resulting in clusters of clonal offshoots (Gräser et al., 2006). The human host is exceptional in lacking a fur, and thus the fungus becomes invasive upon skin contact. This has led to repeated adaptations from zoophilic species with T. equinum originally associated with horse as an example (Kandemir et al., 2020). Several clones in the T. mentagrophytes complex, an originally zoophilic species of small mammals (de Hoog et al., 2017; Kupsch et al., 2019), are now commonly found on humans (Chowdhary et al., 2019; Nenoff et al., 2019a; Das et al., 2020). Clones with mutations in the ribosomal internal transcribed spacers have been assigned numbers (Singh et al., 2019). T. interdigitale, causing chronic, prevalently pedal infections, has been hypothesized to be one of these clones.
Since almost a decade, dermatology in India has experienced a novel driver of dermatophyte evolution due to the overuse of over-the-counter antifungal drugs by the general public (Ebert et al., 2020). An emerging novel species has been named T. indotineae (Kano et al., 2020). This clone shows frequent resistance to terbinafine (>1 μg/ml), which is the most commonly used antifungal to treat dermatophyte infections in India (Kong et al., 2021). In addition, the clone is significantly more virulent than T. interdigitale in the same species complex, causing severe outbreaks (Singh et al., 2019), and is already spreading globally through physical contact and travel (Kano et al., 2020; Jabet et al., 2022; Posso-De Los Rios et al., 2022). Recognition of this clone is therefore clinically significant for proper patient management and public health.
Classically, laboratory diagnosis of dermatophytosis is based on microscopy of strains grown in vitro (Kobylak et al., 2015). Routine fluorescence-microscopy of KOH-digested clinical specimens demonstrating fungal elements is rapid, but unable to differentiate between dermatophytes and non-dermatophyte filamentous fungi (Haghani et al., 2013). Sequencing of rDNA internal transcribed spacer (ITS) in few-days-old cultures is currently the gold standard for dermatophyte identification, despite relatively limited polymorphism (Makimura et al., 1999; Deng et al., 2015). For limited sets of the most common species, a commercial non-culture, molecular assay, DermaGenius 2.0 multiplex real-time PCR kit (Pathonostics, Maastricht, The Netherlands) are available. This tool provides rapid detection of superficial fungal infections of nail, hair, and skin samples and readily differentiates species of Trichophyton, Microsporum and Epidermophyton in addition to Candida albicans (Ndiaye et al., 2022). The kit lacks a probe for T. indotineae, a resistant and hypervirulent dermatophytes species (Singh et al., 2021). The DermaGenius® Resistance Multiplex real-time PCR, is another kit used for detection of terbinafine-resistant T. indotineae strains, but does not recognize susceptible T. indotineae strains (Singh et al., 2021). Other economical molecular methods are available that have not yet been applied to dermatophytes. Loop-mediated isothermal amplification (LAMP) is a rapid assay enabling DNA amplification at constant temperature (Watanabe et al., 2019). Matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-ToF MS) is another routine diagnostic technique for the identification of microorganisms in clinical microbiology laboratories, such as T. rubrum (Wattal et al., 2017; Shaw et al., 2021). Several studies have used MALDI-ToF MS to identify dermatophytes of the T. mentagrophytes complex (Packeu et al., 2013; Calderaro et al., 2014; Shaw et al., 2021). However, recognition of T. indotineae has not been enabled; the reference spectra has not been linked to the species in the database (Calderaro et al., 2014). In this study we determined the performance of AUToF MS 1000 in differentiating the various subspecies by using the profiles of MALDI-ToF MS for hierarchical cluster analysis (HCA) and single-peak analysis.
The expansion of the T. mentagrophytes complex with T. indotineae as a clonal species is of recent date (Tang et al., 2021). T. mentagrophytes, T. interdigitale and T. indotineae are very similar, differing by only a few SNPs in ITS region (Singh et al., 2019; Kano et al., 2020). In order to rapidly and accurately identify the classical species T. mentagrophytes and T. interdigitale and the novelty T. indotineae, current diagnostic assays have to be updated. The present study evaluates several the methods and discusses the taxonomic approach in anthropophilic species complexes.
Materials and methods
Strains and identification
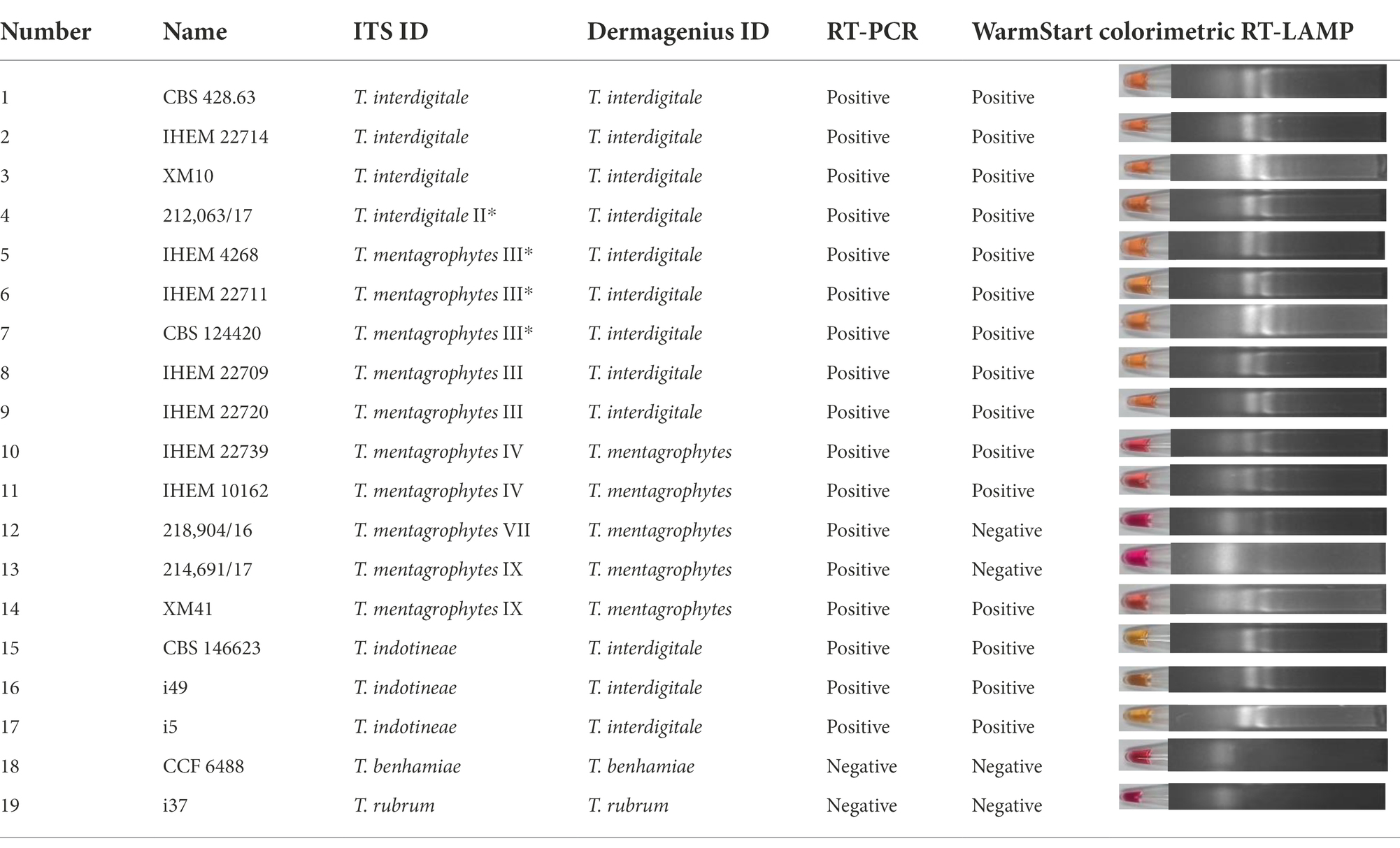
Reference strains were obtained from the Belgian Coordinated Collections of Microorganisms, Scientific Institute of Public Health (BCCM/IHEM, Brussels, Belgium), the Centraalbureau voor Schimmelcultures (CBS, housed at Westerdijk Fungal Biodiversity Institute, Utrecht, Netherlands). Metadata of all 88 strains used in the study are shown in Supplementary Table S1. Strains were cultured on Sabouraud’s Glucose Agar (SGA; Oxoid, Hampshire, UK) for 1–2 weeks at 28°C. DNA extraction and ITS rDNA sequencing according to Arentshorst et al. (2012); Tang et al. (2021). Sequences were blasted in GenBank and 100% identity was taken as species identification. Determination of genotypes were based on ITS sequencing according to Nenoff et al., 2019b and including IHEM 4268 (type of T. mentagrophytes) and CBS 428.63 (type of T. interdigitale) in the comparison. Clone VIII is now known to be identical to the type strain CBS 146623 of T. indotineae (Tang et al., 2021). Forty-nine clinical samples were collected to study DermaGenius (Table 1). Besides, a total of nineteen strains (one to three strains from each genotype) were selected to study DermaGenius, Real-time PCR, and LAMP (Table 2). 81 strains were selected to be studied with MALDI-ToF MS.
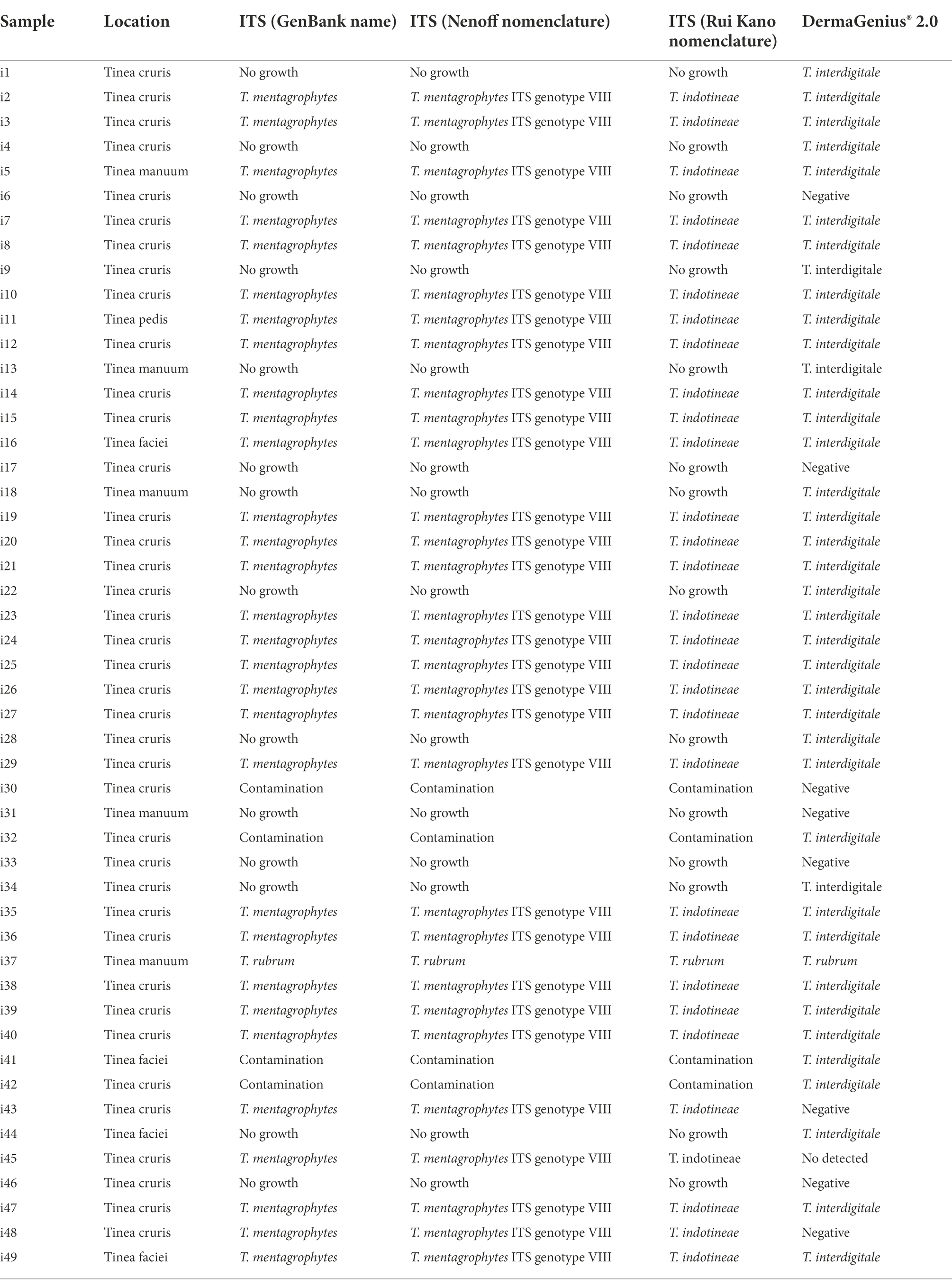
Table 1. Direct culture ITS sequencing and DermaGenius 2.0 PCR results obtained from 49 skin/hair specimens from India.
Dermagenius® 2.0
Forty-nine clinical samples (skin scrapings and hair) from symptomatic patients in India were collected by R. Thakur (Table 1). Each specimen was divided into three parts: the first one for SGA culture, the second for culture on Taplin agar (Oxoid, Munich, Germany), and the third for direct non-culture diagnostics with DermaGenius. Materials were cultured on agar for 1–2 weeks at 28°C (Tang et al., 2021). For DNA extraction, hair and skin scrapings were added to sterile 1.5 mL tubes and extracted with glass beads (Sigma G9143, St. Louis, USA) using the PathoNostics Extraction Kit according to the manufacturer’s instructions. Quality of DNA was tested based on methods described by Arentshorst et al. (2012).
Besides, nineteen strains were selected out of 88 strains to perform DermaGenius testing (Table 2). The DermaGenius® 2.0 complete multiplex real-time PCR (PathoNostics, Maastricht, The Netherlands) was performed according to manufacturer’s instructions. Five μL of DNA extract was added to the PCR mix and a LightCycler 480 II (Roche, Mannheim, Germany) was used for amplification and melting curve analysis. Positive and negative controls were included in each PCR run. Data analysis was performed using the 2nd-derivative and Tm-calling function of the LightCycler 480 II software (v1.5.1.62 SP2).
Real-time PCR
Nineteen strains were selected to represent the described genotypes and species based on the ITS gene region; this marker was used to design the primers and probes shown in Supplementary Table S2.1 DNA of reference strains representing all genotypes of the T. mentagrophytes complex was purified, and amplified by real-time PCR using a T. indotineae-specific probe. Real-time PCR reactions were carried out in 20 μL volumes containing 0.5 μL of 10 μM forward primer, 0.5 μL of 10 μM reverse primer, 0.5 μL probe, 0.5 μL of 40 to 100 ng/μL DNA, and 10 μL LightCycler® 480 Probe Master. The instrument used was a LightCycler® 480 II (Kobylak et al., 2015). Reaction conditions were as follows: 95°C for 10 min, followed by 50 cycles at 95°C for 15 s, 68°C or 60°C for 1 min, with an extension cycle of 40°C for 15 s. The curves indicate positive and the straight lines indicate negative samples (Supplementary Figure S3). Water was used as negative control. CBS 146623 was used as positive control.
Lamp
WarmStart colorimetric Loop-mediated isothermal amplification (LAMP) was applied to detect T. interdigitale, T. mentagrophytes and T. indotineae, with T. benhamiae and T. rubrum as negative controls, through isothermal amplification of the ITS gene. The same nineteen strains as for Real-time PCR were used to represent genotypes (Table 1). The ITS type-specific LAMP primer sets were designed by using the software NEB LAMP Primer Design Tool,2 consisting of two outer (F3, B3), two inner (FIP, BIP) primers, and two loop primers (LF, LB). Used primers are shown in Supplementary Table S2. Reactions were performed according to WarmStart Colorimetric LAMP 2× Master Mix (New England Biolabs, Ipswich, UK) protocol (Dao Thi et al., 2020; Daskou et al., 2021). Samples were incubated for 15 min and 30 min at 65°C in a heating block. Color change was visible by visual observation directly upon removal from the incubation temperature. As additional verification, amplification products were analyzed by 2% agarose gel electrophoresis and visualized under an UV transilluminator. Water was used as negative control. CBS 146623 was used as positive control. Red and yellow indicate negative and positive results, respectively.
MALDI-ToF MS
The data include T. mentagrophytes (n = 23), T. interdigitale (n = 19), T. indotineae (n = 33), T. benhamiae (n = 3), T. quinckeanum (n = 3). MALDI-ToF MS was performed by the formic acid extraction method according to the manufacturer’s instruction (AUToF MS1000, Autobio, Zheng Zhou, China) with minor modifications. All chemical reagents used were of LC–MS grade. Briefly, dermatophytes isolates were cultured on SGA for 7 days at 28°C. After growth, the sample was collected in a 1.5 mL centrifuge tube containing 0.5 mL 75% ethanol. After mixing, the sample was centrifuged at 12,000 × g for 3 min, and the supernatant was discarded. After drying of the residue at 37°C, 40 μL of lysis solution 1 (containing formic acid) was added. Subsequently, 1 μL of supernatant was transferred on a target plate (Autobio, Zheng Zhou, China) after mixing, and dried naturally in a bio-safety cabinet. Afterward, 1 μL of matrix solution was added on the above dried supernatant, and dried again at room temperature. Each strain was prepared on eight MALDI target positions in parallel. For each strain, a mass spectrum was generated and integrated to a sum spectrum using AUToF MS1000. Finally, five spectra were selected from each species for better spectra handling and visualization. MALDI-tree was built up by software inside of AUTO MS1000 with hierarchical cluster analysis.
Results
Clinical samples
DermaGenius 2.0 found the majority of clinical samples from India to be positive for dermatophytes (n = 40/49, 81.63%). Of these, 39 were identified as T. interdigitale, and one as T. rubrum (Table 1; Supplementary Figure S1). Fungal cultures on SGA and Taplin agar yielded 31 (63.26%) samples positive for dermatophytes (Table 1). DermaGenius identified the India samples as T. interdigitale. Comparison of ITS sequences generated from the strains (Table 1; Supplementary Figure S1), 30 isolates matched with T. mentagrophytes ITS-genotype VIII according to Nenoff et al. (2019a) and with CBS 146623, the type strain of T. indotineae.
Nineteen strains from the culture collection representing each genotype using the ITS classification of Nenoff et al. (2019a) were tested by DermaGenius (Supplementary Figure S2). DermaGenius correctly recognized genotypes IV, VII, and IX as belonging to T. mentagrophytes, and all strains of T. interdigitale. However, genotype III and III* of T. mentagrophytes were recognized as T. interdigitale (Table 2). T. indotineae, not present in the DermaGenius database, was identified as T. interdigitale.
Real-time PCR
The assays were positive for all genotypes, while controls (T. rubrum and T. benhamiae) remained negative (Table 2; Supplementary Figure S3). The ITS variable positions used for primer and probe design did not allow distinction between genotypes, which was maximally 1 bp difference between entities.
Warmstart colorimetric RT-LAMP
WarmStart Colorimetric RT-LAMP assay was first executed to determine the incubation time range. The experiment result shows the optimal incubation time is 10 to 20 min. Moreover, an incubation period of 30 min leads to false-positive results with non-template-containing samples as well as with non-T. mentagrophytes complex samples. When samples were incubated at 65°C for 15 min, those matching with T. indotineae, T. mentagrophytes III*, III, and IV, and T. interdigitale were positive (Table 2; Supplementary Figure S4). T. benhamiae, T. rubrum, T. mentagrophytes genotype VII, one strain of T. mentagrophytes genotype IX, and water were negative (Table 2; Supplementary Figure S4). The colorimetric assay evaluated by visual observation was confirmed on the gel (Supplementary Figure S4). However, the protocol did not allow to separate all genotypes unambiguously.
MALDI-ToF mass spectrometry
Representative isolates of each species from the T. mentagrophytes clade were analyzed using MALDI-ToF MS. High-quality (peak rich) MALDI spectra samples were selected to build the MALDI-ToF MS tree. A clustering MALDI-ToF MS analysis results for 81 isolates are shown in Supplementary Table S1; Figure 1. All T. benhamiae and T. quinckeanum strains clustered in their clades and 96.97% (32/33) of T. indotineae strains clustered also together. Six T. interdigitale strains, in the upper single clade, were almost isolated from cases with onychomycosis. The remaining strains, mostly T. mentagrophytes, did not form regularly clusters. CBS 146623, CBS 428.63, CBS 124421 were selected to represent the spectra of T. indotineae, T. interdigitale, and T. mentagrophytes, respectively. In the mass range between approximately 4,000 to 5,000 m/z (as a representative example), the MALDI-ToF MS of CBS 146623, CBS 428.63, and CBS 124421 were very similar, and cannot be differentiated. However, several specific peaks could be found for analyzed taxa in the mass range of approximately between 2,000 to 4,000 and 6,000 to 8,000 m/z (Supplementary Figure S5). The most variable mass range of approximately 2,000 to 4,000 m/z is shown in Supplementary Figure S5. T. indotineae and T. mentagrophytes showed peaks at 2,206 m/z. T. interdigitale and T. indotineae showed peaks at 2,610 to 2,680 m/z. T. interdigitale and T. mentagrophytes showed high peaks at 3,810 to 3,830 and 7,800 to 8,000 m/z, but T. indotineae showed very low peaks at these ranges (Figure 2).
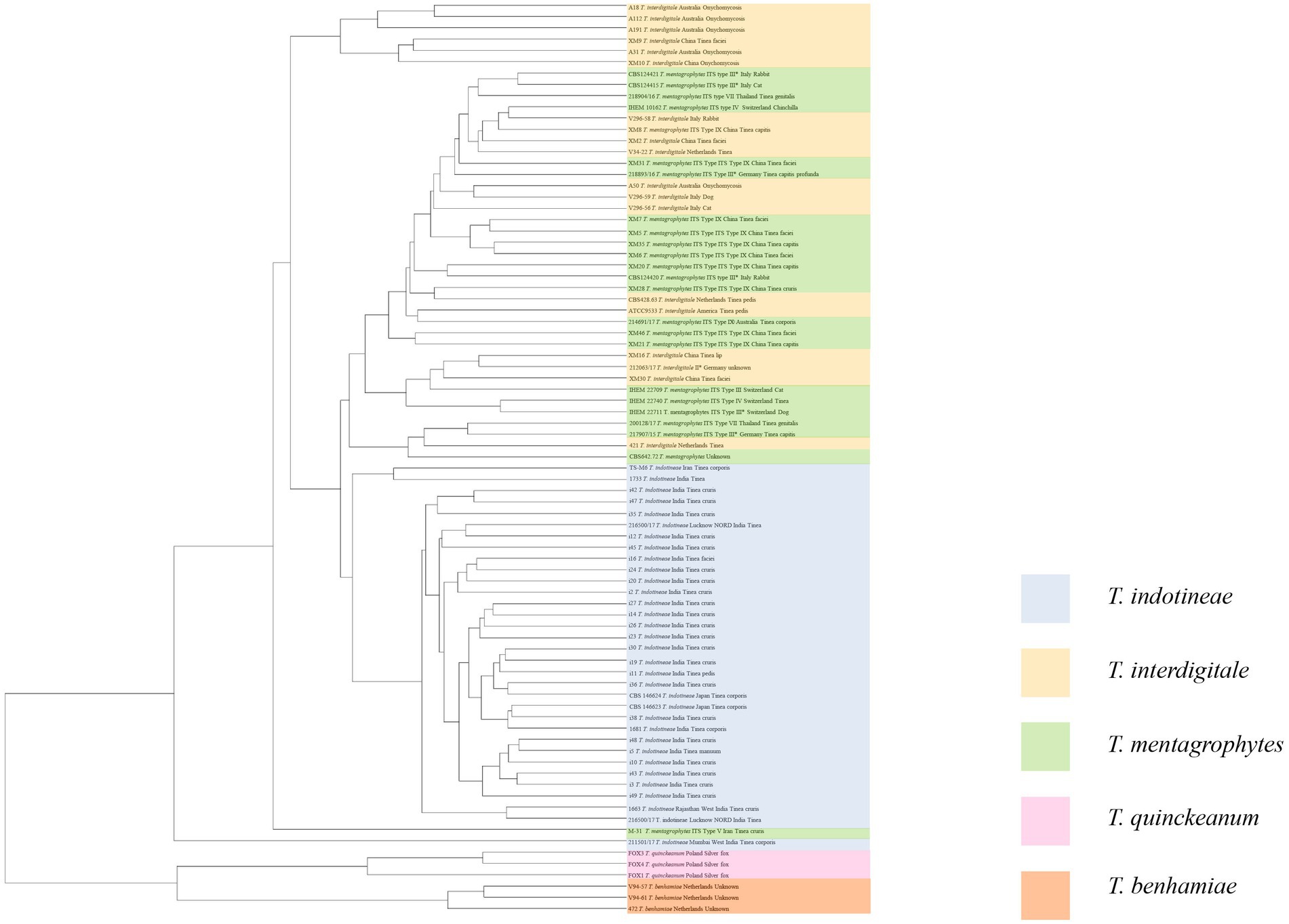
Figure 1. Distance dendrogram of MALDI-ToF MS analysis of T. mentagrophytes species complex. T. benhamiae is outgroup.
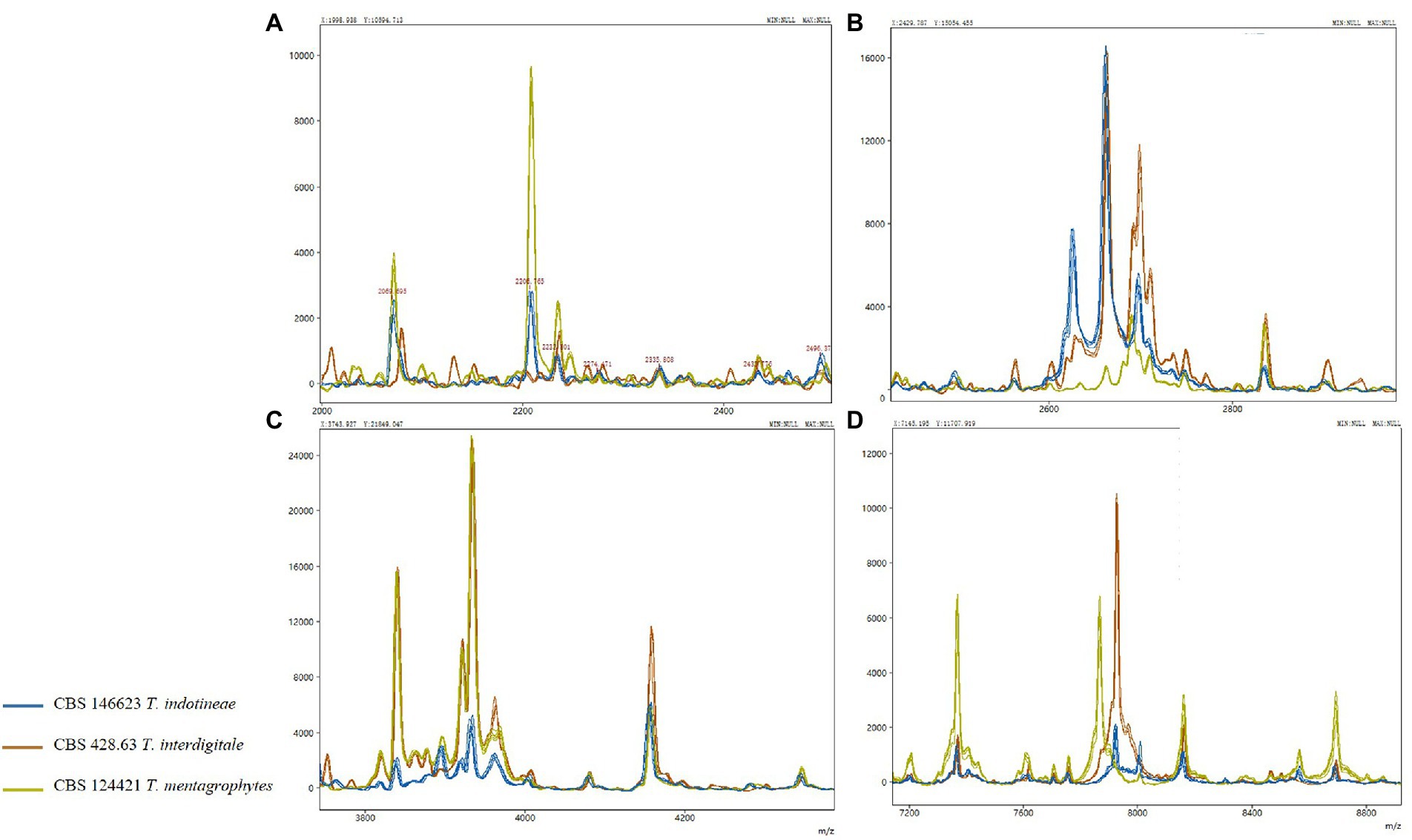
Figure 2. MALDI-ToF MS in the T. mentagrophytes species complex; only variable regions are shown in (A–D).
Discussion
Dermatophytes infections have long been regarded as curable, relatively insignificant esthetic problems. The severe infections by, e.g., T. schoenleinii have practically disappeared by changes in life style and hygienic measures (Prasanna et al., 2016), and today a wide panel of effective antifungals is available without prescription. However, a new problem has emerged in South Asia and spread to other continents with outbreaks of dermatophyte skin diseases showing a remarkably virulence and resistance to the most commonly used antifungal, terbinafine (Singh et al., 2019; Kong et al., 2021). Several species of dermatophytes are involved in this new problem, including T. rubrum (Shankarnarayan et al., 2020) and particularly species of the T. mentagrophytes complex (Singh et al., 2019). Inappropriate use of antifungal creams with corticosteroids has been hypothesized to be a main reason for this emergence (Verma et al., 2021).
T. indotineae, an emerging dermatophyte in India, is a multidrug-resistant taxonomic novelty, regarded as a clonal offshoot in T. mentagrophytes complex (Singh et al., 2019; Kong et al., 2021; Tang et al., 2021). The main objective of the present study was to evaluate the efficacy of diagnostic methods for rapid detection of this clinically relevant dermatophyte. The DermaGenius® 2.0 multiplex real-time PCR assay is a fast molecular diagnostic method that identifies several dermatophytes directly in nail, hair and skin samples within 3 h (Singh et al., 2021). Compared to traditional methods, such as culture, the DermaGenius kit proved to have a higher detection rate (DermaGenius vs. culture; Table 1). Several samples that did not show growth or yielded contaminants on Taplin agar / SGA were positive for dermatophytes with DermaGenius. However, T. indotineae has not yet been included in the kit. The T. mentagrophytes complex comprises three entities, which are in our dataset, in addition to the ancestral sexually interacting species T. mentagrophytes (comprising ITS genotypes III, III*, IV, VII and IX), two clonal offshoots which have been denominated T. interdigitale (also comprising ITS genotype II) and T. indotineae (ITS genotype VIII) (Nenoff et al., 2019b; Tang et al., 2021); the denominations are after Nenoff et al., 2019a. Since T. indotineae is a new name for T. mentagrophytes genotype VIII, existing databases require updating. DermaGenius made a bipartition in identifying most genotypes (including the T. mentagrophytes type strain IHEM 4268), as T. interdigitale (type strain CBS 428.63), while IV, VII and IX were identified as T. mentagrophytes. The tripartition T. indotineae / T. interdigitale / T. mentagrophytes of Tang et al., 2021 was primarily based on multilocus sequence data (TEF1 and HMG) supplemented with phenotypic markers, matching with minute ITS differences. The DermaGenius probes are based on ITS, but their exact sequence is unknown. Using MEGA v7.0 to align the sequences, we found potential primer sites to separate T. mentagrophytes genotypes IV, VII, and IX from T. interdigitale, T. indotineae, and T. mentagrophytes genotypes III and III* (Supplementary Figure S6). Most doubtful group is III/III*, which variously is classified in T. interdigitale or T. mentagrophytes. Since the latter is the latest a valid species name for this entity, the other databases require updating.
A potential area within ITS for primers design for the simultaneous distinction of all three species sensu stricto (Singh et al., 2019; Tang et al., 2021) just a single SNP of T. indotineae is different from T. mentagrophytes and T. interdigitale (Supplementary Table S2; Supplementary Figure S6). Unfortunately, the number of characteristic SNPs proved insufficient for RT-PCR; in addition, this site was invariable between outgroups T. rubrum and T. benhamiae. A more consistent distinction was observed in the HMG gene, which is the prevalent mating type of T. indotineae (Tang et al., 2021). However, the alpha-box mating type did not reveal a usable difference, and some strains only have one mating type gene (Singh et al., 2019). The ITS region in general did not exhibit difference to design probes and primers.
A similar problem was encountered with other molecular methods tested. In comparison to conventional PCR and real-time PCR, WarmStart® Colorimetric LAMP assay is faster and simpler. Published data showed that the assay does not require expensive special equipment such as a thermal cycler, positive samples being determined by a color change from pink to yellow within 30 min of incubation at 65°C. For recording simple mobile phone cameras can be used (Dao Thi et al., 2020; Reynés et al., 2021). It utilizes four oligonucleotide primers to recognize six different regions of the target gene, while two additional primers, LF and LB, are also incorporated in order to accelerate the amplification reaction and enhance the specificity (Tumino et al., 2020). In this study, colorimetric LAMP was positive with T. indotineae, T. mentagrophytes ITS genotype III*, III, IV and T. interdigitale, but is unable to detect all genotypes in the T. mentagrophytes species complex.
T. indotineae is a member of the T. mentagrophytes complex, an originally zoophilic species which loss ability for sexual reproduction due to domestication of host animals (Tang et al., 2021). The species now mainly consists of a cluster of clonal offshoots, and has a skewed mating type distribution with a preponderance of MAT1-2 (HMG gene). Only two of these clones are as yet clinically significant: the classical species T. interdigitale, mostly causing human pedal infections, and the novel taxon T. indotineae. The latter species shows reasonable specificity (96.97%) with MALDI-ToF MS, but T. interdigitale, which possibly resides on the human host already for prolonged periods, is barely different from other clones, the most reliable approach being ITS sequencing. It may be questioned whether taxonomic distinction of every clone is meaningful for reasons other than epidemiology. For clinical practice, direct analysis of genes which confer resistance may be a good way forward (Burmester et al., 2022).
In the course of evolution, Dermatophytes (family Arthrodermataceae in the order Onygenales) show a trend of adaptation to vertebrate hosts. Ancestral life cycles involve production of elaborate sexual fruiting bodies in the natural environmental and distribution of clones via the fur of terrestrial animals. These species, known as geophiles, have low infective abilities. Zoophilic species are prevalently associated with domesticated animals, have less soil contact and are carried longer in animal fur, as ‘clonal offshoots’(Gräser et al., 2006). A last and most recent adaptation is to the human host who is devoid of fur, and thus superficial infection of the skin takes place rather than asymptomatic colonization of fur. These species tend to adapt to particular body sites and lose sexual reproduction (Metin and Heitman, 2017; Kosanke et al., 2018). The evolution of the family Arthrodermataceae has been estimated to have taken about 37 million year (Kandemir et al., 2021), but Tang et al., 2021 suggested that similar adaptations may proceed rapidly after domestication. Indeed, we observe anthropophilic dermatophytes in several species such as: T. concentricum close to T. benhamiae (Čmoková et al., 2020), T. tonsurans close to T. equinum (Kandemir et al., 2020), and Microsporum ferrugineum close to M. canis (de Hoog et al., 2017; Kosanke et al., 2018). As a result, the human host carries a larger number of specifically adapted dermatophytes than any other mammal. In the T. rubrum complex, no sexual reproduction has been observed. It may be hypothesized that this complex is associated with humans for longer periods, which has led to complete loss of sexuality and divergently adapted clones, such as T. rubrum on glabrous skin and T. violaceum, T. soudanense on the scalp. The evolutionary trend over the entire family can thus be summarized as sexuality with clonal offshoots in terrestrial species (Figure 3A), gradual loss of sexuality with longer transmission periods of clonal offshoots in zoophilic species (Figure 3B), and complete loss of sexuality with specialization of some clones surviving on the human host (Figure 3C). Unisexual reproduction is considered a good strategy for short-term survival and population expansion. There is only limited genetic diversity that might be generated by aneuploidy or chromosomal translocations, which might improve the fitness of the progeny without disturbing a well-adapted genotype and phenotype, while the occurrence of sexual reproduction may enhance its fitness by the introduced limited genetic diversity (Feretzaki and Heitman, 2013; Metin and Heitman, 2017; Kosanke et al., 2018). Sex is more suitable for long-term survival and adaptability to an ever-changing environment (Drenth et al., 2019).
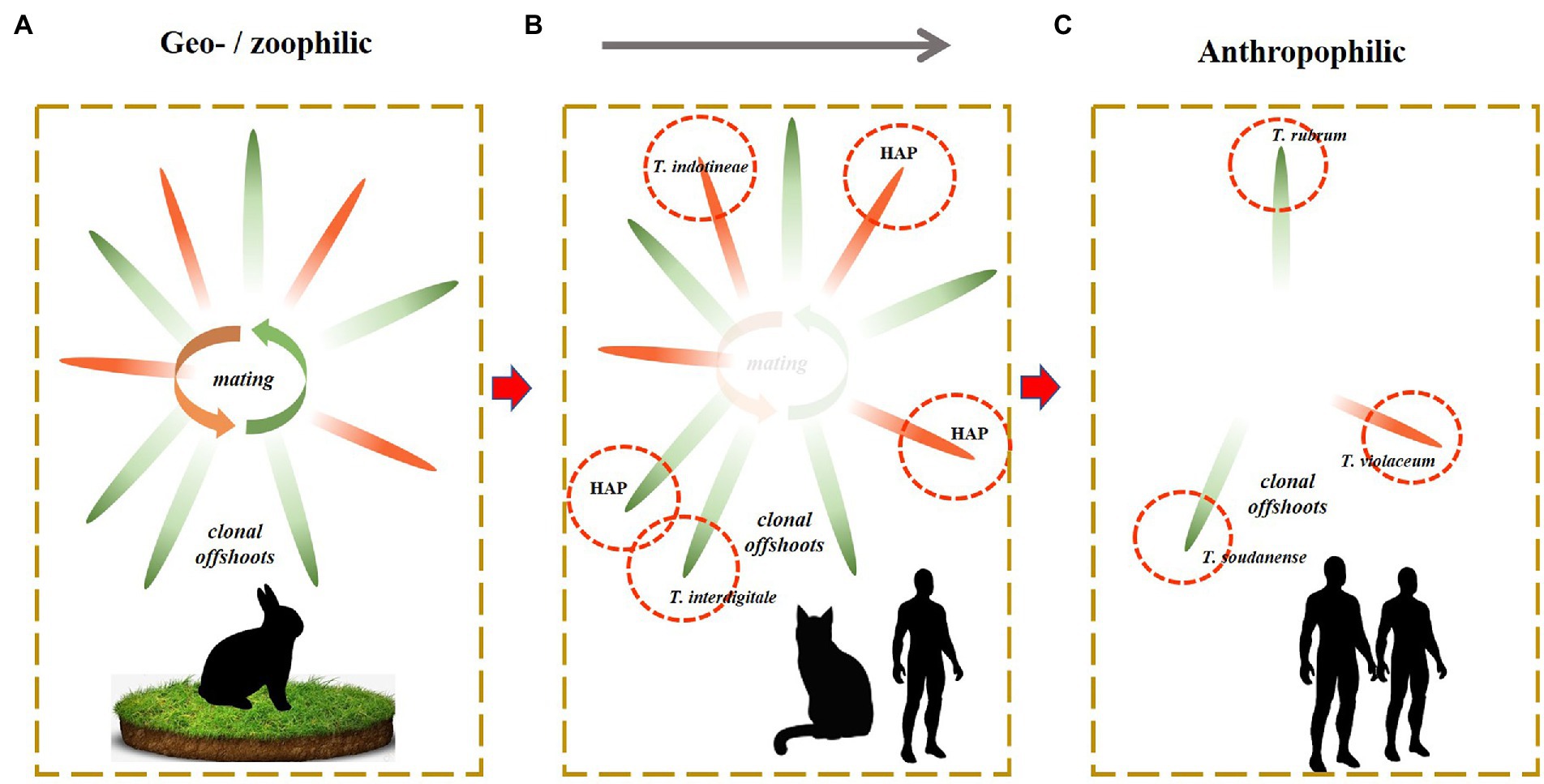
Figure 3. Sexuality with clonal offshoots in terrestrial species (A). Gradual loss of sexuality with longer transmission periods of clonal offshoots in zoophilic species (B). Complete loss of sexuality with specialization of some clones surviving on the human host (C).
In the present paper, we described the intermediate situation (Figure 3B), with decreased sexual reproduction and the presence of numerous clones. Various authors (Nenoff et al., 2019a; Taghipour et al., 2019; Pashootan et al., 2022) distinguished 19 genotypes within the T. mentagrophytes complex, based on ITS sequence diversity. The zoophilic species tend to comprise a plethora of genotypes, as is also observed, e.g., in the T. benhamiae group (Čmoková et al., 2020). With the identification of geophilic dermatophytes, which interact sexually, the genotypes are usually disregarded as just causing some intraspecific variability (Čmoková et al., 2020). In the strictly anthropophilic species of the T. rubrum complex, clones are distinguished because of their ecological adaptation and clinical significance (Su et al., 2019), rather than based on their molecular distance. We recommend to distinguish species of the intermediate group (Figure 3B) only when they show an appreciable degree of adaptation and thus are clinically significant. Clinical relevance is effectuated evolution and is thus also biologically relevant. We advocate to regard the molecular diversity of the T. mentagrophytes complex other than the clones T. indotineae and T. interdigitale as variation within a single species. The lineages within these entities may be epidemiological relevant and can be numbered as haplotypes, rather than attributing formal taxonomic species names.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding authors.
Author contributions
CT, SA, SD, and YK: study conception and design. RT and JZ: sample collection. CT and LZ: data collection. CT, LZ, SA, and JZ: analysis and interpretation of results. CT: draft manuscript preparation. All authors reviewed the results and approved the final version of the manuscript.
Funding
CT was supported by China Scholarship Council (number 201808520089). This work was supported by the 111 Project (D20009); National Natural Science Foundation of China (NSFC; no. 32060034); International Science and Technology Cooperation Base of Guizhou Province [(2020)4101]; Major Science and Technology Plan Project of China National Tobacco Corporation [110202101048 (LS-08)]; Guizhou High-level Innovative Talents Project (qiankeherencai-GCC[2022]036-1) and Talent Base Project of Guizhou Province, China [FCJD2018-22].
Acknowledgments
The authors thank Yinggai Song (Beijing, China) for valuable comments on the manuscript. We thank Hein van der Lee and Marlou Tehupeiory-Kooreman (Nijmegen, The Netherlands) for technical assistance. We are indebted to Ashutosh Singh (Delhi, India) for providing genomic data.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.960190/full#supplementary-material
Footnotes
References
Ameen, M. (2010). Epidemiology of superficial fungal infections. Clin. Dermatol. 28, 197–201. doi: 10.1016/j.clindermatol.2009.12.005
Arentshorst, M., Ram, A. F. J., and Meyer, V. (2012). Using non-homologous end-joining-deficient strains for functional gene analyses in filamentous fungi. Plant Fungal Pathogens 835, 133–150. doi: 10.1007/978-1-61779-501-5_9
Burmester, A., Hipler, U. C., Elsner, P., and Wiegand, C. (2022). Point mutations in the squalene epoxidase erg1 and sterol 14-α demethylase erg11 gene of T indotineae isolates indicate that the resistant mutant strains evolved independently. Mycoses 65, 97–102. doi: 10.1111/myc.13393
Calderaro, A., Motta, F., Montecchini, S., Gorrini, C., Piccolo, G., Piergianni, M., et al. (2014). Identification of Dermatophyte species after implementation of the in-house MALDI-TOF MS database. Int. J. Mol. Sci. 15, 16012–16024. doi: 10.3390/ijms150916012
Celestrino, G. A., Verrinder Veasey, J., Benard, G., and Sousa, M. G. T. (2021). Host immune responses in dermatophytes infection. Mycoses 64, 477–483. doi: 10.1111/myc.13246
Chowdhary, A., Singh, A., Singh, P. K., Khurana, A., and Meis, J. F. (2019). Perspectives on misidentification of Trichophyton interdigitale/Trichophyton mentagrophytes using internal transcribed spacer region sequencing: urgent need to update the sequence database. Mycoses 62, 11–15. doi: 10.1111/myc.12865
Čmoková, A., Kolařík, M., Dobiáš, R., Hoyer, L. L., Janouškovcová, H., Kano, R., et al. (2020). Resolving the taxonomy of emerging zoonotic pathogens in the Trichophyton benhamiae complex. Fungal Divers. 104, 333–387. doi: 10.1007/s13225-020-00465-3
Dao Thi, V. L., Herbst, K., Boerner, K., Meurer, M., Kremer, L. P. M., Kirrmaier, D., et al. (2020). A colorimetric RT-LAMP assay and LAMP-sequencing for detecting SARS-CoV-2 RNA in clinical samples. Sci. Transl. Med. 12:eabc7075. doi: 10.1126/SCITRANSLMED.ABC7075
Das, S., De, A., Saha, R., Sharma, N., Khemka, M., and Singh, S. (2020). The current indian epidemic of dermatophytosis: A study on causative agents and sensitivity patterns. Indian J. Dermatol. 65, 118–122. doi: 10.4103/ijd.IJD_203_19
Daskou, M., Dimitriou, T. G., Alexopoulou, D. S., Tsakogiannis, D., Amoutzias, G. D., Mossialos, D., et al. (2021). WarmStart colorimetric RT-LAMP for the rapid, sensitive and specific detection of enteroviruses A–D targeting the 5′UTR region. J. Appl. Microbiol. 130, 292–301. doi: 10.1111/jam.14770
Deng, S., Bulmer, G. S., Summerbell, R. C., de Hoog, G. S., Hui, Y., and Gräser, Y. (2008). Changes in frequency of agents of tinea capitis in school children from Western China suggest slow migration rates in dermatophytes. Med Mycol. 46, 421–427. doi: 10.1080/13693780701883730
Deng, S., Zhou, Z., de Hoog, G. S., Wang, X., Abliz, P., Sun, J., et al. (2015). Evaluation of two molecular techniques for rapid detection of the main dermatophytic agents of tinea capitis. Br. J. Dermatol. 173, 1494–1500. doi: 10.1111/bjd.14156
de Hoog, G. S., Dukik, K., Monod, M., Packeu, A., Stubbe, D., Hendrickx, M., et al. (2017). Toward a novel multilocus phylogenetic taxonomy for the Dermatophytes. Mycopathologia 182, 5–31. doi: 10.1007/s11046-016-0073-9
Drenth, A., McTaggart, A. R., and Wingfield, B. D. (2019). Fungal clones win the battle, but recombination wins the war. IMA Fungus. 10, 18–16. doi: 10.1186/s43008-019-0020-8
Drouot, S., Mignon, B., Fratti, M., Roosje, P., and Monod, M. (2009). Pets as the main source of two zoonotic species of the Trichophyton mentagrophytes complex in Switzerland, Arthroderma vanbreuseghemii and Arthroderma benhamiae. Vet. Dermatol. 20, 13–18. doi: 10.1111/j.1365-3164.2008.00691.x
Ebert, A., Monod, M., Salamin, K., Burmester, A., Uhrlaß, S., Wiegand, C., et al. (2020). Alarming India-wide phenomenon of antifungal resistance in dermatophytes: A multicentre study. Mycoses 63, 717–728. doi: 10.1111/myc.13091
Feretzaki, M., and Heitman, J. (2013). Unisexual reproduction drives evolution of eukaryotic microbial pathogens. PLoS Pathog. 9:e1003674. doi: 10.1371/journal.ppat.1003674
Gräser, Y., De Hoog, S., and Summerbell, R. C. (2006). Dermatophytes: recognizing species of clonal fungi. Med. Mycol. 44, 199–209. doi: 10.1080/13693780600606810
Haghani, I., Shokohi, T., Hajheidari, Z., Khalilian, A., and Aghili, S. R. (2013). Comparison of diagnostic methods in the evaluation of Onychomycosis. Mycopathologia 175, 315–321. doi: 10.1007/s11046-013-9620-9
Hameed, K., Riaz Ch, F., Nawaz, M. A., Naqvi, S. M. S., Gräser, Y., Kupsch, C., et al. (2017). Trichophyton verrucosum infection in livestock in the Chitral district of Pakistan. J. Infect. Dev. 11, 326–333. doi: 10.3855/jidc.7925
Havlickova, B., Czaika, V. A., and Friedrich, M. (2008). Epidemiological trends in skin mycoses worldwide. Mycoses 51, 2–15. doi: 10.1111/j.1439-0507.2008.01606.x
Jabet, A., Brun, S., Normand, A. C., Imbert, S., Akhoundi, M., Dannaoui, E., et al. (2022). Extensive Dermatophytosis caused by terbinafine resistant Trichophyton indotineae, France. Emerg. Infect. Dis. 28, 229–233. doi: 10.3201/eid2801.210883
Kandemir, H., Dukik, K., Hagen, F., Ilkit, M., Gräser, Y., and de Hoog, G. S. (2020). Polyphasic discrimination of Trichophyton tonsurans and T. equinum from humans and horses. Mycopathologia 185, 113–122. doi: 10.1007/s11046-019-00344-9
Kandemir, H., Dukik, K., de Melo Teixeira, M., Stielow, J. B., Delma, F. Z., Al-Hatmi, A., et al. (2021). Phylogenetic and ecological reevaluation of the order Onygenales. Fungal Divers. 1–72. doi: 10.21203/rs.3.rs-1049506/v1
Kano, R., Kimura, U., Kakurai, M., Hiruma, J., Kamata, H., Suga, Y., et al. (2020). Trichophyton indotineae sp. nov.: A new highly terbinafine-resistant Anthropophilic Dermatophyte species. Mycopathologia 185, 947–958. doi: 10.1007/s11046-020-00455-8
Kobylak, N., Bykowska, B., Nowicki, R., and Brillowska-Dąbrowska, A. (2015). Real-time PCR approach in dermatophyte detection and Trichophyton rubrum identification. Acta Biochim. 62, 119–122. doi: 10.18388/abp.2014_864
Kong, X., Tang, C., Singh, A., Ahmed, S. A., al-Hatmi, A. M. S., Chowdhary, A., et al. (2021). Antifungal susceptibility and mutations in the squalene epoxidase gene in dermatophytes of the Trichophyton mentagrophytes species complex. Antimicrob. Agents Chemotherapy. 65:e0005621. doi: 10.1128/AAC.00056-21
Kosanke, S., Hamann, L., Kupsch, C., Moreno Garcia, S., Chopra, A., and Gräser, Y. (2018). Unequal distribution of the mating type (MAT) locus idiomorphs in dermatophyte species. Fungal Genet. Biol. 118, 45–53. doi: 10.1016/j.fgb.2018.07.003
Kupsch, C., Czaika, V. A., Deutsch, C., and Gräser, Y. (2019). Trichophyton mentagrophytes – a new genotype of zoophilic dermatophyte causes sexually transmitted infections. J. Dtsch. Dermatol. Ges. 17, 493–501. doi: 10.1111/ddg.13776
Makimura, K., Tamura, Y., Mochizuki, T., Hasegawa, A., Tajiri, Y., Hanazawa, R., et al. (1999). Phylogenetic classification and species identification of dermatophyte strains based on DNA sequences of nuclear ribosomal internal transcribed spacer 1 regions. J. Clin. Microbiol. 37, 920–924. doi: 10.1128/jcm.37.4.920-924.1999
Metin, B., and Heitman, J. (2017). Sexual reproduction in Dermatophytes. Mycopathologia 182, 45–55. doi: 10.1007/s11046-016-0072-x
Morrell, J., and Stratman, E. (2011). Primary care and specialty care delays in diagnosing Trichophyton verrucosum infection related to cattle exposure. J. Agromedicine. 16, 244–250. doi: 10.1080/1059924X.2011.605715
Ndiaye, M., Sacheli, R., Diongue, K., Adjetey, C., Darfouf, R., Seck, M. C., et al. (2022). Evaluation of the multiplex real-time PCR DermaGenius® assay for the detection of dermatophytes in hair samples from Senegal. J. Fungi. 8:11. doi: 10.3390/jof8010011
Nenoff, P., Verma, S. B., Vasani, R., Burmester, A., Hipler, U. C., Wittig, F., et al. (2019a). The current Indian epidemic of superficial dermatophytosis due to Trichophyton mentagrophytes—A molecular study. Mycoses 62, 336–356. doi: 10.1111/myc.12878
Nenoff, P., Verma, S. B., Uhrlaß, S., Burmester, A., and Gräser, Y. (2019b). A clarion call for preventing taxonomical errors of dermatophytes using the example of the novel Trichophyton mentagrophytes genotype VIII uniformly isolated in the Indian epidemic of superficial dermatophytosis. Mycoses 62, 6–10. doi: 10.1111/myc.12848
Packeu, A., Hendrickx, M., Beguin, H., Martiny, D., Vandenberg, O., and Detandt, M. (2013). Identification of the Trichophyton mentagrophytes complex species using MALDI-ToF mass spectrometry. Med. Mycol. 51, 580–585. doi: 10.3109/13693786.2013.770605
Pashootan, N., Shams-Ghahfarokhi, M., Chaichi Nusrati, A., Salehi, Z., Asmar, M., and Razzaghi-Abyaneh, M. (2022). Phylogeny, antifungal susceptibility, and point mutations of SQLE gene in major pathogenic Dermatophytes isolated from clinical dermatophytosis. Front. Cellular Inf. Microbiol. 12:851769. doi: 10.3389/fcimb.2022.851769
Posso-De Los Rios, C. J., Tadros, E., Summerbell, R. C., and Scott, J. A. (2022). Terbinafine resistant Trichophyton indotineae isolated in patients with superficial Dermatophyte infection in Canadian patients. J. Cutan. Med. Surg. 120347542210778. doi: 10.1177/12034754221077891
Prasanna, S., Kandasamy, M., Srinivasan, G., and Karthika, J. (2016). Trichophyton schoenleinii: An Unusual Cause of Tinea corporis. International J. Allied Med. Sci. Clin. Res. 4:1.
Reynés, B., Serra, F., and Palou, A. (2021). Rapid visual detection of SARS-CoV-2 by colorimetric loop-mediated isothermal amplification. BioTechniques. 70, 218–225. doi: 10.2144/BTN-2020-0159
Sardana, K., Gupta, A., and Mathachan, S. R. (2021). Immunopathogenesis of Dermatophytoses and factors leading to recalcitrant infections. Indian Dermatol. Online J. 12, 389–399. doi: 10.4103/IDOJ.IDOJ_503_20
Shankarnarayan, S. A., Shaw, D., Sharma, A., Chakrabarti, A., Dogra, S., Kumaran, M. S., et al. (2020). Rapid detection of terbinafine resistance in Trichophyton species by amplified refractory mutation system-polymerase chain reaction. Sci. Reports. 10, 1297–1296. doi: 10.1038/s41598-020-58187-0
Shaw, D., Ghosh, A. K., Paul, S., Singh, S., Chakrabarti, A., Kaur, H., et al. (2021). Matrix-assisted laser desorption/ionisation-time of flight mass spectrometry: Protocol standardisation, comparison and database expansion for faster and reliable identification of dermatophytes. Mycoses 64, 926–935. doi: 10.1111/myc.13285
Singh, A., Masih, A., Monroy-Nieto, J., Singh, P. K., Bowers, J., Travis, J., et al. (2019). A unique multidrug-resistant clonal Trichophyton population distinct from Trichophyton mentagrophytes/Trichophyton interdigitale complex causing an ongoing alarming dermatophytosis outbreak in India: genomic insights and resistance profile. Fungal Genet. Biol. 133:103266. doi: 10.1016/j.fgb.2019.103266
Singh, A., Singh, P., Dingemans, G., Meis, J. F., and Chowdhary, A. (2021). Evaluation of DermaGenius® resistance real-time polymerase chain reaction for rapid detection of terbinafine-resistant Trichophyton species. Mycoses 64, 721–726. doi: 10.1111/myc.13271
Su, H., Packeu, A., Ahmed, S. A., al-Hatmi, A. M. S., Blechert, O., İlkit, M., et al. (2019). Species distinction in the Trichophyton rubrum complex. J. Clin. Microbiol. 57, 1–14. doi: 10.1128/JCM.00352-19
Taghipour, S., Pchelin, I. M., Zarei Mahmoudabadi, A., Ansari, S., Katiraee, F., Rafiei, A., et al. (2019). Trichophyton mentagrophytes and T. interdigitale genotypes are associated with particular geographic areas and clinical manifestations. Mycoses 62, 1084–1091. doi: 10.1111/myc.12993
Tang, C., Kong, X., Ahmed, S. A., Thakur, R., Chowdhary, A., Nenoff, P., et al. (2021). Taxonomy of the Trichophyton mentagrophytes/T. interdigitale species complex harboring the highly virulent, multiresistant genotype T. indotineae. Mycopathologia. 186, 315–326. doi: 10.1007/s11046-021-00544-2
Tumino, S., Tolone, M., Parco, A., Puleio, R., Arcoleo, G., Manno, C., et al. (2020). Validation of loop-mediated isothermal amplification (LAMP) field tool for rapid and sensitive diagnosis of contagious Agalactia in small ruminants. Animals 10:509. doi: 10.3390/ANI10030509
Verma, S. B., Panda, S., Nenoff, P., Singal, A., Rudramurthy, S. M., Uhrlass, S., et al. (2021). The unprecedented epidemic-like scenario of dermatophytosis in India: III. Antifungal resistance and treatment options. Indian J. Dermatol. Venereol. Leprol. 87, 468–482. doi: 10.25259/IJDVL_303_20
Watanabe, S., Okubo, A., Miyajima, Y., Satoh, K., and Makimura, K. (2019). Specific detection of Trichophyton rubrum and Trichophyton interdigitale based on loop-mediated isothermal amplification (LAMP) from onychomycosis specimens. J. Dermatol. 46, 1179–1183. doi: 10.1111/1346-8138.15092
Wattal, C., Oberoi, J. K., Goel, N., Raveendran, R., and Khanna, S. (2017). Matrix-assisted laser desorption ionization time of flight mass spectrometry (MALDI-TOF MS) for rapid identification of micro-organisms in the routine clinical microbiology laboratory. Eur. J. Clin. Microbiol. Infect. Dis. 36, 807–812. doi: 10.1007/s10096-016-2864-9
Keywords: Trichophyton indotineae, Trichophyton mentagrophytes species complex, resistance, taxonomy, evolution, detection, diagnosis, Maldi-ToF
Citation: Tang C, Ahmed SA, Deng S, Zhang L, Zoll J, Al-Hatmi AMS, Meis JF, Thakur R, Kang Y and de Hoog GS (2022) Detection of emerging genotypes in Trichophyton mentagrophytes species complex: A proposal for handling biodiversity in dermatophytes. Front. Microbiol. 13:960190. doi: 10.3389/fmicb.2022.960190
Edited by:
Apichai Tuanyok, University of Florida, United StatesReviewed by:
Mariusz Dyląg, University of Wrocław, PolandMaíra Pompeu Martins, University of São Paulo, Brazil
Yvonne Gräser, Charité Universitätsmedizin Berlin, Germany
Copyright © 2022 Tang, Ahmed, Deng, Zhang, Zoll, Al-Hatmi, Meis, Thakur, Kang and de Hoog. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yingqian Kang, kangyingqian@gmc.edu.cn; G. Sybren de Hoog, sybren.dehoog@radboudumc.nl
 Chao Tang
Chao Tang Sarah A. Ahmed
Sarah A. Ahmed Shuwen Deng
Shuwen Deng Lu Zhang
Lu Zhang Jan Zoll
Jan Zoll Abdullah M. S. Al-Hatmi
Abdullah M. S. Al-Hatmi Jacques F. Meis
Jacques F. Meis Rameshwari Thakur
Rameshwari Thakur Yingqian Kang1*
Yingqian Kang1* G. Sybren de Hoog
G. Sybren de Hoog