A common statement on anthropogenic hybridization of the European wildcat (Felis silvestris)
- 1Department of Evolutionary Biology and Environmental Studies, University of Zurich, Zurich, Switzerland
- 2Wildtier Schweiz, Zurich, Switzerland
- 3CIBIO, Centro de Investigação em Biodiversidade e Recursos Genéticos, InBIO Laboratório Associado/BIOPOLIS Program in Genomics, Biodiversity and Land Plannin, Universidade do Porto, Porto, Portugal
- 4Department of Biology, Faculty of Sciences University of Porto, Porto, Portugal
- 5School of Biological Sciences, University of Bristol, Bristol, United Kingdom
- 6Department of Genetics and Evolution, University of Geneva, Geneva, Switzerland
- 7Institute of Genetics and Genomics in Geneva (IGE3), University of Geneva, Geneva, Switzerland
- 8CNRS, Laboratoire de Biométrie et Biologie Evolutive, Université Claude Bernard Lyon 1, Villeurbanne, France
- 9Department of National Park Monitoring and Animal Management, Bavarian Forest National Park, Grafenau, Germany
- 10Chair of Wildlife Ecology and Management, Albert Ludwigs University Freiburg, Freiburg, Germany
- 11Faculty of Applied Ecology, Agricultural Sciences and Biotechnology, Inland Norway University of Applied Sciences, Koppang, Norway
- 12Unit for Conservation Genetics (BIO-CGE), Italian Institute for Environmental Protection and Research (ISPRA), Ozzano dell Emilia, Italy
- 13Centre for Wildlife Genetics & Conservation Genetics Section, Senckenberg Research Institute and Natural History Museum Frankfurt, Gelnhausen, Germany
- 14LOEWE Centre for Translational Biodiversity Genomics, Frankfurt am Main, Germany
- 15Royal Zoological Society of Scotland (RZSS), WildGenes Laboratory, Edinburgh, United Kingdom
- 16EBM, Biological Station of Mértola, Mértola, Portugal
Preserving natural genetic diversity and ecological function of wild species is a central goal in conservation biology. As such, anthropogenic hybridization is considered a threat to wild populations, as it can lead to changes in the genetic makeup of wild species and even to the extinction of wild genomes. In European wildcats, the genetic and ecological impacts of gene flow from domestic cats are mostly unknown at the species scale. However, in small and isolated populations, it is known to include genetic swamping of wild genomes. In this context, it is crucial to better understand the dynamics of hybridization across the species range, to inform and implement management measures that maintain the genetic diversity and integrity of the European wildcat. In the present paper, we aim to provide an overview of the current scientific understanding of anthropogenic hybridization in European wildcats, to clarify important aspects regarding the evaluation of hybridization given the available methodologies, and to propose guidelines for management and research priorities.
Introduction
We use the taxonomy of Kitchener et al. (2017) when referring to Felis silvestris, Felis lybica and Felis catus. Felis silvestris describes European forest wildcats, Felis lybica includes steppe and bush wildcats with a wide distribution across Mediterranean Islands, Africa and Asia. Felis catus refers to domestic cats. We include indoor–outdoor, free-ranging and feral cats (following the definitions given by Crowley et al., 2020) but exclude cats that do not have the potential to hybridize with Felis silvestris, i.e., all cats living outside the distribution range of wildcats, as well as indoor cats (following the definition given by Crowley et al., 2020). The term hybridization is used in a broad sense, including introgression, i.e., gene flow beyond the first generation.
Hybridization as a conservation issue
Protecting wild species against hybridization with their domestic relatives is an important conservation issue. Keeping and promoting biodiversity, including the genetic diversity of wild species, is a legal obligation anchored in national and international laws and conventions, including the Bern Convention and the European Habitat Directive (92/43 CE) and the Kunming-Montreal global biodiversity framework (COP15). Direct and indirect human impact has altered species distributions worldwide and brought previously isolated taxa into close contact. The breakdown of isolating mechanisms between species may have various consequences, including biodiversity loss resulting from genetic erosion and the merging of distinct evolutionary lineages (Allendorf et al., 2001; Mallet, 2005; Abbott et al., 2016; Quilodrán et al., 2020a, Trouwborst and Somsen, 2019; Barton and Hewitt, 1985; Barton, 2001; Harrison and Larson, 2014; Adavoudi and Pilot, 2021). Anthropogenic hybridization is defined as the admixture between two taxa as a result of human action, including but not limited to species introduction, habitat disturbance, or escape and feralization of domestic species (McFarlane and Pemberton, 2019), and is considered an important threat to biodiversity preservation. In particular, hybridization between wild species and their domestic counterparts promotes the spread and the frequency of traits favored during domestication that can be selectively disadvantageous in natural settings, negatively impacting the fitness of wild individuals (McGinnity et al., 2003; Tymchuk et al., 2007; Muhlfeld et al., 2009; Gering et al., 2019). Moreover, domestication may induce relaxation of natural selection factors, where some traits and their associated genes, usually existing at very low frequencies in nature, have higher frequencies in the domestic forms. Thus, anthropogenic hybridization will disturb the genetic background of natural populations, and in some cases induce outbreeding depression (Frankham et al., 2011). Though positive effects are also possible, particularly when associated with adaptive introgression of beneficial alleles (Feulner et al., 2013; Grossen et al., 2014), the increasing prevalence of so-called “domestic genes”, i.e. genes positively selected in domestication, are likely to be maladaptive in nature. Therefore, based on a precautionary principle, we should assume an overall negative effect of anthropogenic hybridization in wildlife and limit its occurrence.
European wildcats, Felis silvestris, can hybridize with domestic cats, Felis catus, which was domesticated from the African wildcat, Felis lybica, to produce fertile offspring (Driscoll et al., 2009; Kitchener et al., 2017). This contrasts with hybridization between wolves and dogs or wild boars and pigs, for example, where the domestic form descends directly from the wild species with which it hybridizes. Indeed, domestic cats are genetically closer to Felis lybica, a species that has evolved in a different ecological and evolutionary context than Felis silvestris. Cat domestication has involved an artificial selection of traits adapted to life in human care that are common to pets, including cranial volume, gut length, seasonal home range, reproductive rate, behavior, and tameness (Daniels et al., 2002; Kitchener et al., 2005; Germain et al., 2008; Klar et al., 2008; Devillard et al., 2014; Grimm, 2014; Montague et al., 2014; Berteselli et al., 2017). Therefore, domestic cat traits are potentially maladaptive in the ecological context of the European wildcat, as shown in other contexts (McGinnity et al., 2003; Araki et al., 2007; Gering et al., 2019). European wildcats are the focus of this article, but hybridization between domestic cats and other native wildcat species across Africa and Eurasia, though poorly studied, is likely to pose a similar risk.
Although maladaptive domestic traits are expected to be selected against in natural settings, domestic animals usually outnumber their wild counterparts so overwhelmingly that these unbalanced population sizes can still lead to a genetic swamping effect, potentially resulting in the genetic extinction of the smaller wild population (Rhymer and Simberloff, 1996; Allendorf et al., 2001; Field et al., 2008). Domestic cats are ubiquitous across the European wildcat range and are expected to be far more numerous than wildcats (in 2021, in the European Union: 78 million pet cats (Statista.com) and roughly 50,000–75,000 wildcats (expert guess, based on population estimates given in the IUCN Red List Assessment). Data available from various countries provide examples of the order of magnitude: in 2013, in Germany, 11.5 million domestic cats (Statista.com) and approx. 5,000–7,000 wildcats (German federal office for nature protection); in 2020, in Switzerland, 1.7 million domestic cats (Statista.com) and approx. 1,000–3,000 wildcats (Swiss federal office for environment). The reproductive barriers between both species are weak: hybridization occurs and results in fertile offspring. We hypothesize that, at least in regions with lower densities of wildcats than domestic cats, the limited number of intraspecies mating partners can lead wildcats to mate with the ubiquitous and more abundant domestic cats (see also discussion in Oliveira et al., 2018 and Tiesmeyer et al., 2020). We should therefore expect high levels of introgression of domestic cat genes into wildcat populations as a result of this demographic unbalance (Rhymer and Simberloff, 1996; Allendorf et al., 2001; Todesco et al., 2016). The Scottish wildcat is an important example of extensive domestic introgression due to recent human-mediated hybridization. In Scotland, the wild population currently resembles a hybrid swarm (Senn et al., 2018; Breitenmoser et al., 2019), resulting from repeated backcrossing and mating between hybrids, creating a genetic continuum between domestic cats and wildcats (Senn et al., 2018; Howard-McCombe et al., 2021). The prevalence of hybrids, which have a mix of parental morphological traits, confounds accurate field identification (and therefore population monitoring) and legal protection for wildcats. Currently, the low number of remaining wildcats, all introgressed to some extent, has led to a critical state where the wildcat population in Scotland is considered no longer viable (Breitenmoser et al., 2019). Modeling of genomic data demonstrates acute hybridization in Scotland is recent and has likely developed in the last ~60 years in response to a population decline and fragmentation. Thus, hybridization in Scotland is likely to be a consequence of the small population size (Howard-McCombe, 2021; Howard-McCombe et al., 2021).
The fitness consequences of wildcat hybridization are poorly understood, but we can speculate on potential ecological consequences. Domestic cats have a wider diet and habitat spectrum than European wildcats (Germain et al., 2009). If this follows the level of introgression, hybridized wildcats may enlarge their prey and habitat spectrum. Altered home-range sizes (Ellington and Murray, 2015) and social systems, resulting in potentially much larger mesopredator biomass per area, may lead to severely increased predation. Such change in the ecological function of the wildcats may negatively impact the ecosystem (Adavoudi and Pilot, 2021). Moreover, introgression may lead to increased tolerance of wildcats for urban areas, driving more frequent contact with domestic cats and thus acceleration of hybridization, and more frequent exposure to other threats, such as diseases, traffic, or poaching. Further, some introgressed domestic genes might improve adaptation to anthropogenic habitats. More studies are needed to address the fitness impacts of hybridization on individuals and populations of European wildcats, including, e.g., reproductive success, survival rate, behavior, and ecological requirements of cats with different degrees of admixture.
Hybridization levels and population dynamics
The admixture rate between wildcats and domestic cats varies substantially across the species range, from almost 100% in Scotland to less than 2% of individuals of recent admixed ancestry in Central Germany. In several Central European countries, the hybridization rate is about 5%, whereas in southern Europe it can reach over 20% (Tiesmeyer et al., 2020; Urzi et al., 2021). While partly reflecting an artefact due to variation in methods to quantify introgression, this regional variation of admixture rate may be explained by local population dynamics. Hybridization may be more frequent in small or fragmented wildcat populations, where low wildcat density and a lack of intraspecific mating partners favor hybridization events (Hubbs, 1955), as is reported for dogs and wolves (Galaverni et al., 2017). Population size determines the relative impact of any hybridization event, which will be generally larger in small populations. In small and fragmented populations, animals are proportionally more often located on fringes (edge effect, Primack, 1993). Therefore, small and isolated populations are especially threatened by hybridization due to stochastic, demographic and adaptive factors, including allele swamping, outbreeding depression, and genomic extinction through assimilation of the wild species by the domestic taxon (Todesco et al., 2016). For many small and declining wildcat populations, levels of hybridization higher than the European average have been reported, e.g., 21% in Mediterranean regions of the Iberian Peninsula (Tiesmeyer et al., 2020; Matias et al., 2022), and in Scotland, where a continuing high rate of hybridization has generated a hybrid swarm effect (Beaumont et al., 2001; Senn et al., 2018; Howard-McCombe, 2021).
Hybridization may be more frequent in the context of range expansion (Currat et al., 2008). This could especially affect wildcats, as they re-colonize previous areas of their range that are now human-dominated landscapes with a large domestic cat presence (Quilodrán et al., 2020b). Re-colonized areas typically show low wildcat population density and scarcity of intraspecific mating partners, resulting in elevated hybridization propensity and rapid introgression (Klopfstein et al., 2006; Excoffier et al., 2009). For example, hybridization in Switzerland is best explained by wildcat range expansion during the last fifty years, rather than by domestic cats invading wildcat habitats (Nussberger et al., 2018; Quilodrán et al., 2019). Recent range expansion has been documented in several other countries (e.g., France: Léger et al., 2008; Netherlands: Janssen et al., 2016; Germany: Steyer et al., 2016; Mueller et al., 2020; northern Italy: Spada et al., 2022). This would be consistent with the conclusion of several studies that domestic introgression into wild populations is a recent phenomenon (Goedbloed et al., 2012; Roed et al., 2014, Howard-McCombe et al., 2021).
The role of domestic cats in hybridization with wildcats
Domestic cats have been widespread in Europe at least since their introduction by the Romans (Faure and Kitchener, 2009; Krajcarz et al., 2020). There is still substantial genetic differentiation between domestic cats and European wildcats, suggesting the presence of reproductive barriers between the two species (Tiesmeyer et al., 2020; Nieto-Blázquez et al., 2022). However, this assumption should not be used as proof of the harmlessness of long-term hybridization between both taxa. Indeed, we are observing a continuous increase in the number of domestic cats and an overall degradation of environmental conditions (e.g., habitat loss, habitat fragmentation, and human disturbance) that might threaten the long-term persistence of the wildcat (Sarmento et al., 2009). Pet cat ownership continues to increase into this century. For example, between 1965 and 2000 the number of pet cats in the UK doubled, from 4 to 8 million (based on the statistics of the UK pet food, www.ukpetfood.org). In Germany the number almost doubled between 2010 and 2019, from 8.2 to 14.8 million (welt-der-katzen.de). Even if many “pet cats” are not living in contact with wildcats and may be neutered, we assume the increased number of “pet cats” can still be used as a proxy for the number of “domestic cats” as we define them in this paper. Even if domestic cats are found more often in vicinity of human settlements and less often in large, forested areas, the preferred wildcat habitat, the domestic cat habitat spectrum is still quite large and also includes wildcat habitats (Berberat, 2021; Nussberger et al., 2023). We assume that an increasing number of domestic cats leads to more domestic cats present in wildcat habitat, thus more encounters and thus more mating events with wildcats. Based on this assumption, increasing domestic cat populations are likely to promote hybridization. In addition, habitat change or disturbance, like forest fragmentation or urbanization of rural areas, may also favor the presence of domestic cats in wildcat habitats and thus further facilitate encounters between both species.
Methods to determine hybridization and their pitfalls
The use of appropriate molecular genotyping methods allows for accurate species discrimination and hybrid level determination. As hybrids can be morphologically indistinguishable from the parental species, hybridization impedes formal identification of wildcats based on phenotypic attributes alone (Hertwig et al., 2009; Krüger et al., 2009; Devillard et al., 2014). A broad correspondence between pelage and genotype has been shown across the hybrid swarm in Scotland (Senn et al., 2018; Howard-McCombe et al., 2021), and this provides a useful approximation for practical management in the absence of a genetic test. However, studies requiring accurate discrimination between wildcats, hybrids and domestic cats, must be based on appropriate genetic analyses – as has also been suggested for other hybridizing species, e.g., wolves (Donfrancesco et al., 2019; Stronen et al., 2022b). Further, one should be aware that conclusions drawn from studies based on an exclusively phenotypical assessment may confound wildcats and their hybrids.
There are several molecular methodologies for assessing hybridization between domestic cats and European wildcats. However, the use of differing molecular approaches can result in artefactual differences in hybridization rates, based on the discrimination power of the applied genetic marker set. Earlier studies (Beaumont et al., 2001; Oliveira et al., 2008a; Oliveira et al., 2008b; Randi, 2008; Hertwig et al., 2009; Say et al., 2012) were mostly based on highly polymorphic markers (microsatellites), initially developed for recognizing population structure in domestic cats, i.e. chosen to have high levels of differentiation (FST-values) within the species (Menotti-Raymond et al., 1999). More recent studies (Nussberger et al., 2014a; Nussberger et al., 2014b; Oliveira et al., 2015; Senn et al., 2018; Tiesmeyer et al., 2020) rely on diagnostic markers (SNPs), specifically designed in domestic cats and European wildcats to increase power in assessing hybridization, i.e. chosen to have low FST-values within the species but very high FST-values between the hybridizing species (Nussberger et al., 2013; Mattucci et al., 2019). Polymorphic microsatellites and diagnostic SNPs can be viewed as complementary molecular approaches. Polymorphic microsatellites are best used for assessing population structure but might also be able to detect early stages of hybridization. A panel of pre-selected diagnostic SNPs offers greater accuracy and discrimination power to identify and quantify hybridization, and higher data comparability between laboratories (Steyer et al., 2018; Harmoinen et al., 2021). The two approaches are correlated, as illustrated by Supplementary Table 2, showing a set of wildcat samples assigned based on polymorphic microsatellites and diagnostic SNPs (see also introduction in Nussberger et al., 2013 and discussion in Steyer et al., 2018). However, the following important points should be considered: (i) in studies using polymorphic markers, cross-checking between polymorphic and diagnostic markers is advisable to make reliable statements about hybridization; (ii) when comparing different genetic studies, it is essential to check that the genetic markers that are used show similar overall discrimination power between the hybridizing species (depending on the number of markers and the level of differentiation, e.g., FST-values). Also, as polymorphic microsatellites are highly sensitive toward population substructure, biased choice of reference populations may lead to false-positive hybrid identifications (Steyer et al., 2016; Stronen et al., 2022a). Ideally, genetic studies that need to be compared with each other should use the same genetic marker sets.
Genetic markers on mitochondrial DNA can also be used as a complementary information in the context of the hybridization, to trace the maternal line and thus the directionality of the hybridization. However, mtDNA alone is not recommended for studying hybridization, since it does not account for any information on the paternal line neither from the nuclear genomic component. In addition, the evolution of mitochondrial genome reflects the evolution of the organelles, more than the evolution of the organisms carrying these mitochondria and cases of mitochondrial introgression can mislead data interpretation (Alves et al., 2008). Biases in the interpretation of hybridization and species divergence due to the use of only the mitochondrial genome have already been reported, for example, polar bears hybridizing with brown bears (Hailer et al., 2012).
The sampling method is another crucial aspect to reliably detect the extent of admixture in a population. We advocate systematic collection of samples, rather than opportunistic sampling, to better represent the local population. We encourage repeating the sampling regularly, as hybridization will likely vary over time, following demographic changes in the wild and domestic populations.
To date, hybridization between European wildcats and domestic cats has primarily been assessed through opportunistic sampling, mostly using roadkill. Such studies give only a snapshot in space and time of the hybridization level in a given population. Opportunistic approaches can be misleading: roadkill surveys usually only collect samples from cats with the typical wildcat phenotype, which a priori exclude many hybrid individuals showing a less typical phenotype (Steyer et al., 2018). On the other hand, cats from remote areas, where roadkill is rare, are underrepresented. Systematic sampling to monitor hybridization should not be limited to core areas of known wildcat distribution, but instead be representative of the species distribution, including its edges where hybrids may be more common. Even though they are necessary to reveal long-term trends, studies based on systematic population sampling over a large area and a longer timeframe are still very scarce (e.g., Léger et al., 2008; Nussberger et al., 2014a).
Further, we urge caution when interpreting hybridization levels. When hybridization levels are below ~10%, they are often qualified as low or insignificant (e.g., Steyer et al., 2016; Beugin et al., 2020). Such interpretations may lead to the premature conclusion that hybridization is harmless, in the sense of being no threat to the long-term persistence of the population or species, and, as such, not a serious concern for wildcat conservation. Delimitation of “low” and “harmless” hybridization levels should not be based on an arbitrary threshold but should instead be based on the amount of admixture that can be tolerated without risking the loss of adaptive wildcat genetic diversity. Currently, there is not enough scientific evidence to estimate a tolerance threshold (and it is likely to vary between populations). Therefore, it is premature to qualify existing hybridization levels as harmless amounts of admixture. Indeed, model projections show that hybridization tends to increase in almost all simulated scenarios, and that even seemingly low rates of hybridization are sufficient to assimilate wildcats to domestic cats over 50% in less than 100 years (Quilodrán et al., 2020b). Recent evidence shows the hybrid swarm effect has developed even more rapidly in Scotland (Howard-McCombe, 2021; Howard-McCombe et al., 2021).
Management implications
Based on a conservationist precautionary principle, we recommend practitioners to consider any hybridization level as potentially harmful, and to monitor the evolution of hybridization rates. This will in turn provide further evidence to estimate the levels of domestic gene flow potentially leading to irreversible genetic diversity loss. We predict that hybridization tolerance levels will be especially low for small and low-density European wildcat populations.
While it is currently unclear to what degree hybridization with domestic cats poses a threat to the European wildcat’s long-term survival and its network of ecological interactions, gene flow from the domestic to the wild species should be minimized as much as possible. This statement is based on (i) a precautionary principle, since negative evolutionary consequences cannot be excluded, and (ii) on a fundamental conservation goal, which is to maintain natural adaptive genetic diversity in wild species. Minimizing gene flow requires a combined effort, involving policymakers (e.g., countrywide or regional governmental agencies in charge of wildlife conservation, as well as politicians), conservation managers, game keepers, hunters, conservation practitioners, and citizens, especially cat owners.
In parallel, we need to acquire more knowledge of many aspects of wildcat biology. Scientists should focus on assessing the impact of introgression from domestic cats, especially regarding fitness consequences. We need a better understanding of the genotype-phenotype map of wildcats and domestic cats to evaluate the impact of any level of hybridization. Fitness effects of hybridization may depend on the genomic region that is introgressed, with some genomic regions having stronger effects than others. More work is also needed to improve our understanding of when and how hybridization events occur and what their consequences are by, e.g., studying the behavior and ecology of wildcats, domestic cats, and hybrids with different levels of admixture – as done by Matias et al. (2022) in a study about variation of genetic integrity across biomes. This knowledge gained by scientific research is essential for defining efficient conservation and management measures (Jackiw et al., 2015). Expertise exchange and joint projects between researchers from the disciplines of population genetics, wildcat ecology, population ecology, eco-evolutionary modeling, veterinary medicine, and captive management should be encouraged, as interdisciplinarity is invaluable to effective implementation of conservation programmes (Stoskopf et al., 2005). Comparing findings from different wildcat populations across Europe will be especially valuable. The Eurowildcat Consortium, a scientific network connecting wildcat researchers across Europe, initiated at a wildcat symposium in Mulhouse in 2013 and formally founded in 2018, was a first step in this direction. Nevertheless, the management still differs widely across European countries. For example, in some countries wildcats are considered as endangered, especially due to high hybridization risk (e.g., Portugal). In other European countries wildcats are game species without a management plan or systematic monitoring, despite high levels of hybridization have been recorded (e.g., Serbia; Urzi et al., 2021).
Based on current knowledge, we recognize the following priorities in conservation management and scientific research concerning hybridization between wildcats and domestic cats (see also Table 1):
● Management measures: Due to the lack of knowledge about their eco-evolutionary consequences, we should prevent hybridization events as far as possible (complete avoidance is not realistic), especially by minimizing the presence of fertile domestic cats in wildcat habitats and promoting healthy wildcat populations.
● Systematic monitoring: Policymakers should implement long-term genetic monitoring programmes with reliable temporal and spatial sampling to assess hybridization at a biologically meaningful local scale. Additionally, demographic trends should be regularly determined (stable, expanding, decreasing populations). Ideally, demographic trends in domestic–feral cats in the wildcat range should also be measured. Long-term data are valuable to understand the spatial and temporal drivers of hybridization. It would allow conservationists to evaluate the success of measures taken to minimize hybridization events.
● Communication with the wider public: Conservation managers should encourage projects to raise awareness of hybridization as a conservation issue and promote responsible pet ownership. Domestic cat owners should be aware of their responsibility to keep pets from damaging the natural environment. Information campaigns describing wildcats (as a separate species from domestic cats) could use the wildcat as an attractive flagship species for conservation, as has been done for example in Germany with the projects “Rettungsnetz Wildkatze” and “Wildkatzensprung” (governmental project of BUND). A well-informed public would increase the acceptance of domestic and stray cat management measures and engagement with wildcat conservation in general.
● Domestic cat management: Policymakers should encourage neutering programmes for owned or semi-owned domestic cats living in or near typical wildcat habitats (e.g., isolated farms in rural areas, far away from highly urbanized regions). This measure aims to minimize successful mating events between the wild and domestic species. Further, mandatory microchipping of domestic cats could give a systematic census of the domestic cats. We recommend controlling feral cat populations by eradication/neutering programmes as far as possible. Due to the emotional attachment to cats, trap–neuter–return (TNR) strategies are generally the best accepted and thus the most realistic management option to minimize successful mating events between wildcats and stray/feral cats. Monitoring should be used to verify the long-term efficacy of this strategy. A further approach to domestic cat management was chosen by the Polish Academy of Sciences, who added Felis catus to the list of “invasive alien species”, emphasizing in this way the threat this species represents to biodiversity (e.g., impact as a predator shown by Kosicki, 2021).
● Managing wildcat habitat: Wildcat habitat recovery should be promoted, by increasing habitat heterogeneity (e.g., less intensive silviculture and agriculture, more deadwood), mitigating roadkill hotspots, avoiding fragmentation or increasing connectivity between wildcat habitats, and promoting habitat characteristics to increase prey species abundance (e.g., hedgerows). Habitat improvement may lead to stable or increasing wildcat populations. Wildcats are predicted to choose native forest and woodland areas, avoid human structures like villages, houses, and roads (Klar et al., 2008), and use well-structured habitats (Jerosch et al., 2010; Jerosch et al., 2017). Conversely, poor quality habitat may lead to wildcat dispersal into human-dominated landscapes, and subsequent hybridization with domestic cats. Matias et al. (2022) have shown that wildcat genetic integrity was generally maximized in regions with a higher forest cover and larger distances to human settlements. Thus, maintaining large, connected native forests and woodlands with low anthropogenic disturbance is pivotal for maintaining large, demographically and genetically stable wildcat populations.
● Limiting human disturbance in wildcat regions: Many different sources of human disturbance negatively affect wildcats, such as increased stress levels due to the presence of intensive tourism (Piñeiro et al., 2012) or mistaken shooting or poaching (even if rare in most countries nowadays). Thus, we expect wilderness areas mostly free from human pressures in whichever form (tourism, urbanization, hunting) to benefit wildcat populations. In healthy, large wildcat populations, intraspecies mating might be favored, thus reducing the probability of hybridization with domestic cats.
● Legal protection against hybridization: The European wildcat is a legally protected species in most European countries. The protection primarily applies to prevent hunting. Nevertheless, legal protection could be extended to include the genetic integrity and diversity of the species. Policymakers might revise legal texts to support measures against the negative effects of hybridization more explicitly. For example, in Switzerland, there is a legal duty to ensure that hybrids that have entered the wild are regulated and do not spread, and where possible, are removed if they threaten native species diversity (Hunting ordinance, SR 922.01, Art. 8bis, al. 5). Legislation concerning the regulation of feral cats as an invasive species might also have the potential for improvement (Trouwborst and Somsen, 2019).
● Communication with hunters: To avoid mistakenly shooting or poaching of wildcats in countries where wildcats are legally protected, hunters should be informed about the presence of the species, its legal status and problems associated with hybridization with domestic cats. They should be advised not to shoot tabby cats. The European wildcat is protected in most European countries, e.g., by the Council Directive on the conservation of natural habitats and of wild fauna and flora (92/43/EEC). In contrast, feral cats can be hunted in some countries, e.g., United Kingdom, Germany, Switzerland, Slovakia or Czechia, which may reduce hybridization, but – in the absence of adequate information campaigns – might also increase mistaken shooting of wildcats.
● Research: We should fully understand hybridization dynamics and the impact of gene flow from domestic cats to European wildcats.
○ What are the fitness consequences of hybridization at the level of individuals, populations, and species? Individuals: promising approaches include systematically quantifying differential survival of kittens with or without recent genetic admixture, or of radio-collared individuals with or without recent admixture, as well as quantifying fecundity or disease prevalence of admixed and non-admixed wildcats. Populations: life history traits, e.g., reproduction, survival and growth rate, of populations with different hybridization levels should be compared to better understand the impact of hybridization at a population level. Species: we should better understand how selection is operating on the flow of domestic cat genes into the wildcat gene pool, which genes are more likely to be introgressed and what are the genotype–phenotype maps of the cat species. Here, genomic approaches will be of great importance.
○ How does behavior, habitat use, and morphology, differ between parental species and hybrids with different levels of admixture? Understanding the behavioral isolating mechanisms and the spatial configuration of wildcats, domestic cats and their hybrids is crucial to understanding (and limiting) hybridization events (Matias et al., 2022). The study of species-specific habitat use may identify optimal areas for measures to mitigate hybridization. The study of morphological traits allows better identification of hybrids in the field, which is crucial for the fast implementation of management measures.
○ How might hybridization rates change in future? Simulated data can be used to project future patterns of hybridization based on past and current hybridization levels (as in Quilodrán et al., 2020b). It would be useful to compare simulation models for several populations with different baselines, in terms of population parameters, like hybridization rate, or effective population size of the parental species. This might be relevant to help policymakers define population-specific thresholds for “acceptable” levels of gene flow of hybridization.
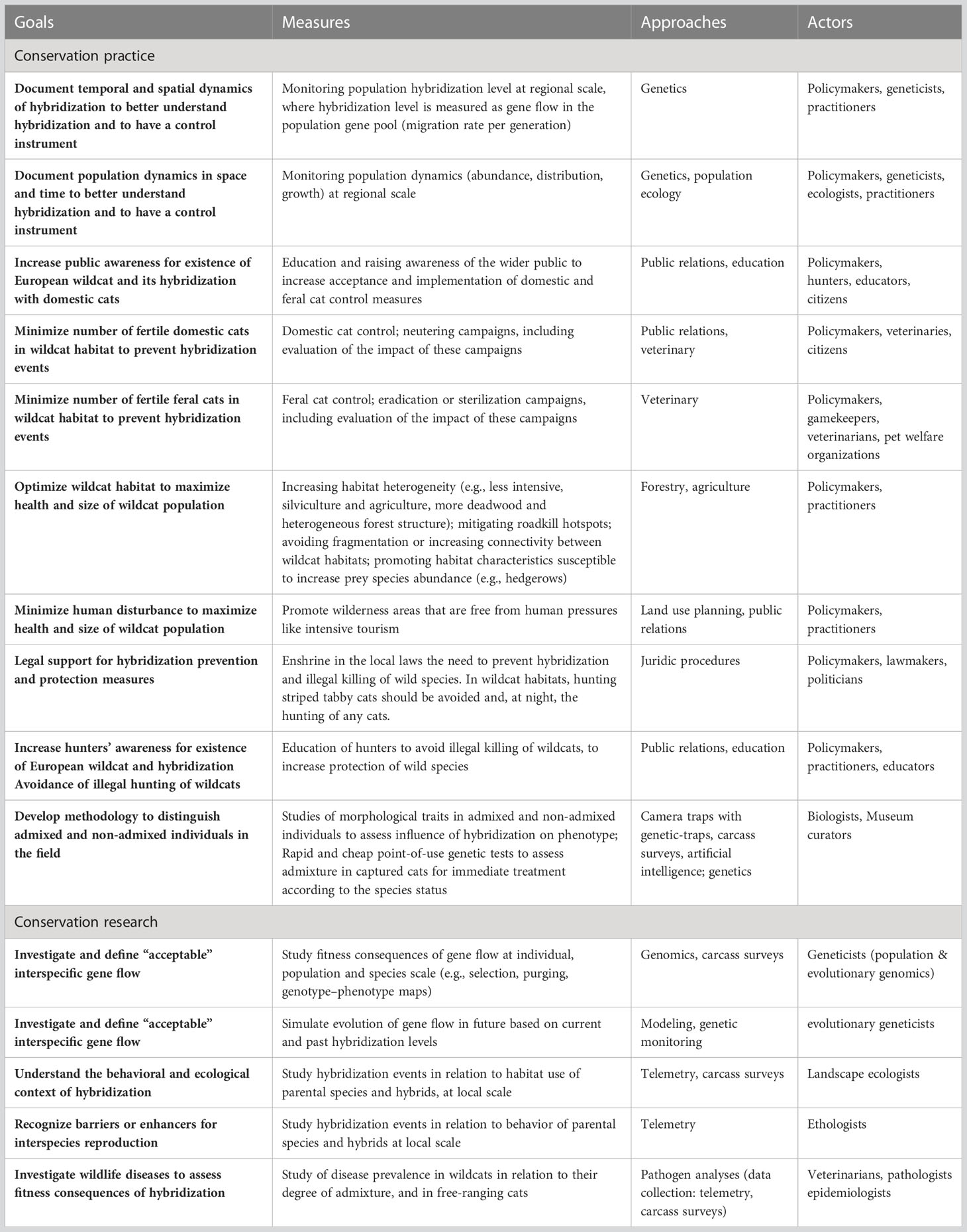
Table 1 Overview of conservation priorities for Felis silvestris regarding its hybridization with Felis catus under the responsibility of policymakers (conservation practice) or researchers (conservation research).
Summary
Hybridization between European wildcats and domestic cats appears to be a modern phenomenon, a result of the breakdown of isolating mechanisms between species previously isolated for thousands of years. Historic habitat loss, persecution and current trends in urbanization, habitat fragmentation and domestic cat ownership suggest hybridization now poses a significant threat to wildcats, especially in small, isolated, or declining populations. We currently have a limited understanding of the main drivers of hybridization, fitness consequences for wildcats or ecosystem function of hybrids. Despite reported cases of “low” levels of hybridization across countries in central Europe, a cautious approach should be taken to managing hybridization. Low levels of introgression can quickly result in genetic swamping, as has already been observed in Scotland, where the wildcat population is now on the verge of extinction. Systems that appear stable may be capable of switching rapidly into a hybrid swarm if further disturbed, be this by human forces (e.g., habitat loss or hunting) or natural events (e.g., prey crashes). Accurate genetic methods now exist to quantify hybridization and introgression, and we strongly encourage their consequent application to monitor wildcat populations periodically. Mitigating hybridization poses complex management questions, requiring the combined effort of multiple stakeholders, including scientists, and engagement of the citizens. However, it is vital to be aware of, and manage, the risks of hybridization to retain this charismatic cat species and preserve its distinct gene pool in Europe.
Author contributions
BN, SB, MB, MC, SD, MH, JH-M, FM, CN, CSQ, HS, and PCA: wrote and reviewed manuscript. EUROWILDCAT Consortium: supported the statement. All authors contributed to the article and approved the submitted version.
Funding
Work co-funded by NORTE-01-0246-FEDER-000063, supported by Norte Portugal Regional Operational Programme (NORTE2020), under the PORTUGAL 2020 Partnership Agreement, through the European Regional Development Fund (ERDF).
Acknowledgments
We thank all the members of the Eurowildcat Consortium that inspired us to write this article and for the productive discussions.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2023.1156387/full#supplementary-material
References
Abbott R., Barton N. H., Good J. M. (2016). Genomics of hybridization and its evolutionary consequences. Mol. Ecol. 25, 2325–2332. doi: 10.1111/mec.13685
Adavoudi R., Pilot M. (2021). Consequences of hybridization in mammals: a systematic review. Genes 13 (1), 50. doi: 10.3390/genes13010050
Allendorf F. W., Leary R. F., Spruell P., Wenburg J. K. (2001). The problems with hybrids: setting conservation guidelines. Trends Ecol. Evol. 16, 613–622. doi: 10.1016/S0169-5347(01)02290-X
Alves P. C., Melo-Ferreira J., Freitas H., Boursot P. (2008). The ubiquitous mountain hare mitochondria: multiple introgressive hybridization in hares, genus lepus. Philos. Trans. R. Soc. B: Biol. Sci. 363.1505, 2831–2839. doi: 10.1098/rstb.2008.0053
Araki H., Cooper B., Blouin M. S. (2007). Genetic effects of captive breeding cause a rapid, cumulative fitness decline in the wild. Science 318, 100–103. doi: 10.1126/science.1145621
Barton N. H. (2001). The role of hybridization in evolution. Mol. Ecol. 10 (3), 551–568. doi: 10.1046/j.1365-294x.2001.01216.x
Barton N. H., Hewitt G. M. (1985). Analysis of hybrid zones. Annu. Rev. Ecol. Systematics. 16, 113–148. doi: 10.1146/annurev.es.16.110185.000553
Beaumont M., Barratt E. M., Gottelli D., Kitchener A. C., Daniels M. J., Pritschard J. K., et al. (2001). Genetic diversity and introgression in the Scottish wildcat. Mol. Ecol. 10, 319–336. doi: 10.1046/j.1365-294x.2001.01196.x
Berberat E. (2021). Analysis of the differences between habitat characteristics of wildcats and domestic cats in potential wildcat habitats in Switzerland (ETH Zurich), 45.
Berteselli G. V., Regaiolli B., Normando S., De Mori B., Zaborra C. A., Spiezio C. (2017). European Wildcat and domestic cat: do they really differ? J. Veterinary Behav. 22, 35–40. doi: 10.1016/j.jveb.2017.09.006
Beugin M. P., Salvador O., Leblanc G., Queney G., Natoli E., Pontier D. (2020). Hybridization between felis silvestris silvestris and felis silvestris catus in two contrasted environments in France. Ecol. Evol. 10, 263–276. doi: 10.1002/ece3.5892
Breitenmoser U., Lanz T., Breitenmoser-Würsten C. (2019). Conservation of the wildcat (Felis silvestris) in Scotland: review of the conservation status and assessment of conservation activities (IUCN SSC Cat Specialist Group).
Crowley S., Cecchetti M., McDonald R. A. (2020). Our wild companions: domestic cats in the anthropocene. Trends Ecol. Evol. 35 (6), 477–483. doi: 10.1016/j.tree.2020.01.008
Currat M., Ruedi M., Petit R. J., Excoffier L. (2008). The hidden side of invasions: massive introgression by local genes. Evolution 62 (8), 1908–1920. doi: 10.1111/j.1558-5646.2008.00413.x
Daniels M. J., Wright T., Bland K. P., Kitchener A. C. (2002). Seasonality and reproduction in wild-living cats in Scotland. Acta theriologica 47 (1), 73–84. doi: 10.1007/BF03193568
Devillard S., Jombart S., Léger F., Pontier D., Say L., Ruette S., et al. (2014). How reliable are morphological and anatomical characters to distinguish European wildcats, domestic cats and their hybrids in France? J. Zoological Systematics Evolutionary Res. 52, 154–162. doi: 10.1111/jzs.12049
Donfrancesco V., Ciucci P., Salvatori V., Benson D., Andersen L. W., Bassi E., et al. (2019). Unravelling the scientific debate on how to address wolf-dog hybridization in Europe. Front. Ecol. Evol. 7. doi: 10.3389/fevo.2019.00175
Driscoll C. A., Macdonald D. W., O’Brien S. J. (2009). From wild animals to domestic pets, an evolutionary view of domestication. Proc. Natl. Acad. Sci. U. S. A. 106, 9971–9978. doi: 10.1073/pnas.0901586106
Ellington E. H., Murray D. L. (2015). Influence of hybridization on animal space use: a case study using coyote range expansion. Oikos 124, 5. doi: 10.1111/oik.01824
Excoffier L., Foll M., Petit R. J. (2009). Genetic consequences of range expansions. Annu. Rev. Ecology Evol. Systematics 40, 481. doi: 10.1146/annurev.ecolsys.39.110707.173414
Faure E., Kitchener A. C. (2009). An archaeological and historical review of the relationships between felids and people. Anthrozoos: A Multidiscip. J. Interact. People Anim. 22, 221–238. doi: 10.2752/175303709X457577
Feulner P. G. D., Gratten J., Kijas J. W., Visscher P. M., Pemberton J. M., Slate J. (2013). Introgression and the fate of domesticated genes in a wild mammal population. Mol. Ecol. 22 (16), 4210–4221. doi: 10.1111/mec.12378
Field D. F., Ayre D. J., Whelan R. J., Young A. G. (2008). Relative frequency of sympatric species influences rates of interspecific hybridization, seed production and seedling performance in the uncommon Eucalyptus aggregata. J. Ecol. 96, 1198–1210. doi: 10.1111/j.1365-2745.2008.01434.x
Frankham R., Ballou J. D., Eldridge M. D. B., Lacy R. C., Ralls K., Dudash M. R., et al. (2011). Predicting the probability of outbreeding depression. Conserv. Biol. 25 (3), 465–475. doi: 10.1111/j.1523-1739.2011.01662.x
Galaverni M., Caniglia R., Pagani L., Fabbri E., Boattini A., Randi E. (2017). Disentangling timing of admixture, patterns of introgression, and phenotypic indicators in a hybridizing wolf population. Mol. Biol. Evol. 34 (9), 2324–2339. doi: 10.1093/molbev/msx169
Gering E., Incorvaia D., Henriksen R., Wright D., Getty T. (2019). Maladaptation in feral and domesticated animals (Wiley-Blackwell).
Germain E., Benhamou S., Poulle M. L. (2008). Spatio-temporal sharing between the European wildcat, the domestic cat and their hybrids. J. Zoology 276 (2), 195–203. doi: 10.1111/j.1469-7998.2008.00479.x
Germain E., Ruette S., Poulle M. L. (2009). Likeness between food habits of European wildcats, domestic cats and their hybrids in France. Mamm. Biol. 74, 412–417. doi: 10.1016/j.mambio.2009.05.008
Goedbloed D. J., Megens H.-J., Van Hooft P., Herrero-Medrano J. M., Lutz W., Alexandri P., et al . (2012): Data from: Genome-wide single nucleotide polymorphism analysis reveals recent genetic introgression from domestic pigs into Northwest European wild boar populations. Dryad. doi: 10.5061/DRYAD.V6F1G
Grimm D. (2014). The genes that turned wildcats into kitty cats. Science 346 (6211), 799. doi: 10.1126/science.346.6211.799
Grossen C., Keller L., Biebach I., Croll D., International Goat Genome Consortium (2014). Introgression from domestic goat generated variation at the major histocompatibility complex of alpine ibex. PloS Genet. 10 (6), e1004438. doi: 10.1371/journal.pgen.1004438
Hailer F., Kutschera V. E., Hallström B. M., Klassert D., Fain S. R., Leonard J. A., et al. (2012). Nuclear genomic sequences reveal that polar bears are an oldand distinct bear lineage. Science 336, 344–347. doi: 10.1126/science.1216424
Harmoinen J., von Thaden A., Aspi J., Kvist L., Cocchiararo B., Jarausch A., et al. (2021). Reliable wolf-dog hybrid detection in Europe using a reduced SNP panel developed for non-invasively collected samples. BMC Genomics 22 (473), 15. doi: 10.1186/s12864-021-07761-5
Harrison R. G., Larson E. L. (2014). Hybridization, introgression, and the nature of species boundaries. J. Heredity 105 (S1), 795–809. doi: 10.1093/jhered/esu033
Hertwig S. T., Jungnickel A., Stepanow S., Böhle U. R., Fischer M. S. (2009). Regionally high rates of hybridization and introgression in German wildcat populations (Felis silvestris, Carnivora, felidae). J. Zool. Syst. Evol. Res. 47, 283–297. doi: 10.1111/j.1439-0469.2009.00536.x
Howard-McCombe J. (2021). Hybridisation and introgression in the Scottish wildcats: implications for conservation (Doctoral dissertation, University of Bristol). doi: 10.1111/mec.16000
Howard-McCombe J., Ward D., Kitchener A., Lawson D., Senn H., Beaumont M. A. (2021). On the use of genome-wide data to model and date the time of anthropogenic hybridization: an example from the Scottish wildcat. Mol. Ecol. 30 (15), 3688–3702.
Hubbs C. L. (1955). Hybridization between fish species in nature. Systematic Zoology 4 (1), 1–20. doi: 10.2307/2411933
Jackiw R. N., Mandil G., Hager H. A. (2015). A framework to guide the conservation of species hybrids based on ethical and ecological considerations. Conserv. Biol. 29 (4), 1040–1051.
Janssen R., Dekker J. J. A., Mulder J. L., Brouns A., Linnartz L. (2016). De wilde katten van het Vijlenerbos. Onderzoek naar het terreingebruik in 2014-2015. Bionet (Stein) / Jasja Dekker Dierecologie (Arnhem) / Bureau Muldernatuurlijk (Groenekan) Report, 57.
Jerosch S., Götz M., Roth M. (2010). Characteristics of diurnal resting sites of theendangered European wildcat (Felis silvestris silvestris): implications for itsconservation. J. Nat. Conserv. 18 (1), 45–54. doi: 10.1016/j.jnc.2009.02.005
Jerosch S., Götz M., Roth M. (2017). Spatial organisation of European wildcats (Felis silvestris silvestris) in an agriculturally dominated landscape. Cent. Europe Mamm. Biol. 82, 8–16. doi: 10.1016/j.mambio.2016.10.003
Kitchener A. C., Breitenmoser-Würsten Ch., Eizirik E., Gentry A., Werdelin L., Wilting A., et al. (2017). “A revised taxonomy of felidae,” in The final report of the cat classification task force of the IUCN/SSC cat specialist group (Cat News), 80.
Kitchener A. C., Yamaguchi N., Ward J. M., Macdonald D. W. (2005). A diagnosis for the Scottish wildcat (Felis silvestris): a tool for conservation action for a critically-endangered felid. Anim. Conserv. 8 (3), 223–237. doi: 10.1017/S1367943005002301
Klar N., Fernandez N., Kramer-Schadt S., Herrmann M., Trinzen M., Büttner I., et al. (2008). Habitat selection models für European wildcat conservation. Biol. Conserv. 141 (1), 308–319. doi: 10.1016/j.biocon.2007.10.004
Klopfstein S., Currat M., Excoffier L. (2006). The fate of mutations surfing on the wave of a range expansion. Mol. Biol. Evol. 23 (3), 482–490. doi: 10.1093/molbev/msj057
Kosicki J. Z. (2021). The impact of feral domestic cats on native bird populations. predictive modelling approach on a country scale. Ecol. Complexity 48, 100964. doi: 10.1016/j.ecocom.2021.100964
Krajcarz M., Krajcarz M. T., Baca M., Baumann C., Wim Van Neer W., Popović D., et al. (2020). Ancestors of domestic cats in neolithic central Europe: isotopic evidence of a synanthropic diet. PNAS 117, 17710–17719. doi: 10.1073/pnas.1918884117
Krüger M., Hertwig S., Jetschke G., Fischer M. S. (2009). Evaluation of anatomical characters and the question of hybridization with domestic cats in the wildcat population of thuringia, Germany. J. Zool. Syst. Evol. Res. 47, 268–282. doi: 10.1111/j.1439-0469.2009.00537.x
Léger F., Stahl P., Ruette S., Wilhelm J.-L. (2008). La répartition du chat forestier en France: évolutions récentes. Faune Sauvage 280, 24–39.
Mallet J. (2005). Hybridization as an invasion of the genome. Trends Ecol. Evol. 20, 229–237. doi: 10.1016/j.tree.2005.02.010
Matias G., Rosalino L. M., Alves P. C., Tiesmeyer A., Nowak C., Ramos L., et al. (2022). Genetic integrity of European wildcats: variation across biomes mandates geographically tailored conservation strategies. Biol. Conserv. 268, 109518. doi: 10.1016/j.biocon.2022.109518
Mattucci F., Galaverni M., Lyons L. A., Alves P. C., Randi E., Velli E., et al. (2019). Genomic approaches to identify hybrids and estimate admixture times in European wildcat populations. Sci. Rep. 9 11612. doi: 10.1038/s41598-019-48002-w
McFarlane E., Pemberton J. (2019). Detecting the true extent of introgression during anthropogenic hybridization. Trends Ecol. Evol. 34 (4), 315–326. doi: 10.1016/j.tree.2018.12.013
McGinnity P., Prodöhl P., Ferguson A., Hynes R., Maoiléidigh Ó, Baker N., et al. (2003). Fitness reduction and potential extinction of wild populations of Atlantic salmon, salmo salar, as a result of interactions with escaped farm salmon. Proc. R. Soc Lond. B 270, 2443–2450. doi: 10.1098/rspb.2003.2520
Menotti-Raymond M., David V. A., Lyons L. A., et al. (1999). A genetic linkage map of microsatellites in the domestic cat (Felis catus). Genomics 57, 9–23. doi: 10.1006/geno.1999.5743
Montague M. J., Li G., Gandolfi B., Khan R., Aken B.L., Searle S.M.J., et al. (2014). Comparative analysis of the domestic cat genome reveals genetic signatures underlying feline biology and domestication. PNAS 111 (48), 17230–17235. doi: 10.1073/pnas.1410083111
Mueller S. A., Reiners T. E., Steyer K., von Thaden A., Tiesmeyer A., Nowak C. (2020). Revealing the origin of wildcat reappearance after presumed long-term absence. Eur. J. Wildlife Res. 66, 94. doi: 10.1007/s10344-020-01433-7
Muhlfeld C. C., Kalinowski S. T., McMahon T. E., Taper M. L., Painter S., Leary R. F., et al. (2009). Hybridization rapidly reduces fitness of a native trout in the wild. Biol. Lett. 5, 328–331. doi: 10.1098/rsbl.2009.0033
Nieto-Blázquez M. E., Schreiber D., Mueller S. A., Koch K., Nowak C., Pfenninger M., et al. (2022). Human impact on the recent population history of the elusive European wildcat inferred from whole genome data. BMC Genomics 23, 709. doi: 10.1186/s12864-022-08930-w
Nussberger B., Currat M., Quilodrán C. S., Ponta N., Keller L. F. (2018). Range expansion as an explanation for introgression in European wildcats. Biol. Conserv. 218, 49–56. doi: 10.1016/j.biocon.2017.12.009
Nussberger B., Greminger M. P., Grossen C., Keller L. F., Wandeler P. (2013). Development of SNP markers identifying European wildcats, domestic cats, and their admixed progeny. Mol. Ecol. Res. 13, 447–460. doi: 10.1111/1755-0998.12075
Nussberger B., Hertwig S. T., Roth T. (2023). Monitoring distribution, density and introgression in European wildcats in Switzerland. Biol. Cons. 281, 110029. doi: 10.1016/j.biocon.2023.110029
Nussberger B., Wandeler P., Camenisch G. (2014b). A SNP chip to detect introgression in wildcats allows accurate genotyping of single hairs. Eur. J. Wildl Res. 60, 405–410. doi: 10.1007/s10344-014-0806-3
Nussberger B., Wandeler P., Weber D., Keller L. F. (2014a). Monitoring introgression in European wildcats in the Swiss jura. Conserv. Genet. 15, 1219–1230. doi: 10.1007/s10592-014-0613-0
Oliveira R., Godinho R., Randi E., Alves P. C. (2008b). Hybridization vs conservation: are domestic cats threatening the genetic integrity of European wildcat (Felis silvestris silvestris) populations in Iberian peninsula? Philos. Trans. R. Society Biol. Sci. 363, 2953–2961. doi: 10.1098/rstb.2008.0052
Oliveira R., Godinho R., Randi E., Ferrand N., Alves P. C. (2008a). Molecular analysis of hybridization between wild and domestic cats (Felis silvestris) in Portugal: implications for conservation. Conserv. Genet. 9, 1–11. doi: 10.1007/s10592-007-9297-z
Oliveira R., Randi E., Mattucci F., Kurushima J. D., Lyons L. A., Alves P. C. (2015). Towards a genome-wide approach for detecting hybrids: informative SNPs to detect introgression between domestic cats and European wildcats (Felis silvestris). Heredity 115, 195–205. doi: 10.1038/hdy.2015.25
Oliveira T., Urra F., López-Martín J. M., Ballesteros-Duperón E., Barea-Azcón J. M., Moléon M., et al. (2018). Females know better: sex-biased habitat selection by the European wildcat. Ecol. Evol. 8, 9464–9477. doi: 10.1002/ece3.4442
Piñeiro A., Barja I., Silván G., Illera J. (2012). Effects of tourist pressure and reproduction on physiological stress response in wildcats: management implications for species conservation. Wildl Res. 39, 532–539. doi: 10.1071/WR10218
Primack R. B. (1993). Essentials of conservation biology (Sunderland, Massachusetts, USA: Sinauer Associates, Inc.), 601.
Quilodrán C. S., Montoya-Burgos J. I., Currat M. (2020a). Harmonizing hybridization dissonance in conservation. Commun. Biol. 3, 391. doi: 10.1038/s42003-020-1116-9
Quilodrán C. S., Nussberger B., Macdonald D. W., Montoya-Burgos J. I., Currat M. (2020b). Projecting introgression from domestic cats into European wildcats in the Swiss jura. Evolutionary Appl. 13 (8), 2101–2112. doi: 10.1111/eva.12968
Quilodrán C. S., Nussberger B., Montoya-Burgos J. I., Currat M. (2019). Hybridization and introgression during density-dependent range expansion: European wildcats as a case study. Evol. (N. Y). 73, 750–761.
Randi E. (2008). Detecting hybridization between wild species and their domesticated relatives. Mol. Ecol. 17, 285–293.
Rhymer J. M., Simberloff D. (1996). Extinction by hybridization and introgression. Annu. Rev. Ecol. Syst. 27, 83–109. doi: 10.1146/annurev.ecolsys.27.1.83
Roed K. H., Bjornstad G., Flagstad O., Haanes H., Hifthammer A. K., Jordhoy P., et al. (2014). Ancient DNA reveals prehistoric habitat fragmentation and recent domestic introgression into native wild reindeer. Conserv. Genet. 15, 1137–1149.
Sarmento P., Cruz J., Eira C., Fonseca C. (2009). Spatial colonization by fera domestic cats felis catus of former wildcat felis silvestris silvestris home ranges. Acta Theriologica 54 (1), 31–38. doi: 10.1007/BF03193135
Say L., Devillard S., Léger F., Pontier D., Ruette S. (2012). Distribution and spatial genetic structure of European wildcat in France. Anim. Conserv. 15, 18–27. doi: 10.1111/j.1469-1795.2011.00478.x
Senn H. V., Ghazali M., Kaden J., Barclay D., Harrower B., Campbell R. D., et al. (2018). Distinguishing the victim from the threat: SNP-based methods reveal the extent of introgressive hybridization between wildcats and domestic cats in Scotland and inform future in situ and ex situ management options for species restoration. Evol. Appl. 12, 399–414. doi: 10.1111/eva.12720
Spada A., Fazzi P., Lucchesi M., Lazzeri L., Mori E., Velli E., Sforzi A., et al, et al. (2022). An update of the Italian distribution of the European wildcat. Hystrix It. J. Mamm. 33 (XII ATIt Congress Supplement), 124.
Steyer K., Kraus R. H. S., Mölich T., Anders O., Cocchiararo B., Frosch C., et al. (2016). Large-Scale genetic census of an elusive carnivore, the European wildcat (Felis s. silvestris). Conserv. Genet. 17, 1183–1199. doi: 10.1007/s10592-016-0853-2
Steyer K., Tiesmeyer A., Munoz-Fuentes V., Nowak C. (2018). Low rates of hybridization between European wildcats and domestic cats in a human-dominated landscape. Ecol. Evol. 8, 2290–2304. doi: 10.1002/ece3.3650
Stoskopf M. K., Beck K., Fazio B. B., Fuller T. K., Gese E. M., Kelly B. T., et al. (2005). From the field: implementing recovery of the red wolf - integrating research scientists and managers. Wildlife Soc. Bull. 33 (3), 1145–1152. doi: 10.2193/0091-7648(2005)33[1145:FTFIRO]2.0.CO;2
Stronen A. V., Aspi J., Caniglia R., Fabbri E., Galaverni M., Godinho R., et al. (2022a). Wolf-dog admixture highlights the need for methodological standards and multidisciplinary cooperation for effective governance of wild x domestic hybrids. Biol. Conserv. 266, 109467. doi: 10.1016/j.biocon.2022.109467
Stronen A. V., Mattucci F., Fabbri E., Galaverni M., Cocchiararo B., Nowak C., et al. (2022b). A reduced SNP panel to trace gene flow across southern European wolf populations and detect hybridization with other canis taxa. Sci. Rep. 12, 4195. doi: 10.1038/s41598-022-08132-0
Tiesmeyer A., Ramos L., Lucas J.M., Steyer K., Alves P.C., Astaras C., et al. (2020). Range-wide patterns of human-mediated hybridization in European wildcats. Conserv. Genet. 21, 247–260. doi: 10.1007/s10592-019-01247-4
Todesco M., Pascual M. A., Owens G. L., Ostevik K. L., Moyers B.T., Hübner S., et al. (2016). Hybridization and extinction. Evol. Appl. 9, 892–908. doi: 10.1111/eva.12367
Trouwborst A., Somsen H. (2019). Domestic cats (Felis catus) and European nature conservation law–applying the EU birds and habitats directives to a significant but neglected threat to wildlife. J. Environ. Law 32 (3), 391–415. doi: 10.1093/jel/eqz035
Tymchuk W. E., Sundstrom L. F., Devlin R. H. (2007). Growth and survival trade-offs and outbreeding depression in rainbow trout (Oncorhynchus mykiss). Evolution 61, 1225–1237. doi: 10.1111/j.1558-5646.2007.00102.x
Keywords: introgression, Felis silvestris, Felis catus, domestic cat, wildlife management
Citation: Nussberger B, Barbosa S, Beaumont M, Currat M, Devillard S, Heurich M, Howard-McCombe J, Mattucci F, Nowak C, Quilodrán CS, Senn H, Alves PC and EUROWILDCAT Consortium (2023) A common statement on anthropogenic hybridization of the European wildcat (Felis silvestris). Front. Ecol. Evol. 11:1156387. doi: 10.3389/fevo.2023.1156387
Received: 01 February 2023; Accepted: 08 June 2023;
Published: 17 July 2023.
Edited by:
Seth Magle, Lincoln Park Zoo, United StatesReviewed by:
Elena Buzan, University of Primorska, SloveniaMukesh Thakur, Zoological Survey of India, India
Copyright © 2023 Nussberger, Barbosa, Beaumont, Currat, Devillard, Heurich, Howard-McCombe, Mattucci, Nowak, Quilodrán, Senn, Alves and EUROWILDCAT Consortium. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Beatrice Nussberger, beatrice.nussberger@ieu.uzh.ch; Paulo Célio Alves, pcalves@fc.up.pt
†The co-author members of EUROWILDCAT Consortium are listed in Supplementary Table S1
 Beatrice Nussberger
Beatrice Nussberger Soraia Barbosa
Soraia Barbosa Mark Beaumont5
Mark Beaumont5  Mathias Currat
Mathias Currat Sébastien Devillard
Sébastien Devillard Marco Heurich
Marco Heurich Jo Howard-McCombe
Jo Howard-McCombe Federica Mattucci
Federica Mattucci Claudio Sebastián Quilodrán
Claudio Sebastián Quilodrán Helen Senn
Helen Senn Paulo Célio Alves
Paulo Célio Alves