- 1Division of Internal Medicine, Department of Animal Medicine, Faculty of Veterinary Medicine, South Valley University, Qena, Egypt
- 2Division of Infectious Diseases, Department of Animal Medicine, Faculty of Veterinary Medicine, Damanhour University, Damanhour, Egypt
- 3Department of Parasitology, Faculty of Veterinary Medicine, Mansoura University, Mansoura, Egypt
- 4Doctor of Veterinary Sciences, Veterinary Clinic, Veterinary Directorate, Qena, Egypt
- 5Department of Biochemistry, Faculty of Veterinary Medicine, South Valley University, Qena, Egypt
- 6Department of Parasitology, Faculty of Veterinary Medicine, South Valley University, Qena, Egypt
- 7Department of Veterinary Medicine, College of Agriculture and Veterinary Medicine, Qassim University, Buraidah, Saudi Arabia
Introduction: Fascioliasis is a parasitic foodborne disease caused by the liver flukes, Fasciola hepatica and F. gigantica. Such parasites cause serious illness in numerous domestic animals and also in humans. Following infection, the parasite secretes a variety of molecules that immediately interact with the host immunity to establish successful infection. These molecules include cathepsin L peptidase 1 (CatL1); the highly investigated diagnostic and vaccine antigens using various animal models. However, a few studies have analyzed the potentials of FhCatL1 as a diagnostic or vaccine antigen using bioinformatic tools and much less for FgCatL1. The present study provides inclusive and exclusive information on the physico-chemical, antigenic and immunogenic properties of F. hepatica cathepsin L1 (FhCatL1) protein using multiple bioinformatic analysis tools and several online web servers. Also, the validation of our employed available online servers was conducted against a huge collection of previously published studies focusing on the properties of FhCatL1as a diagnostic and vaccine antigen.
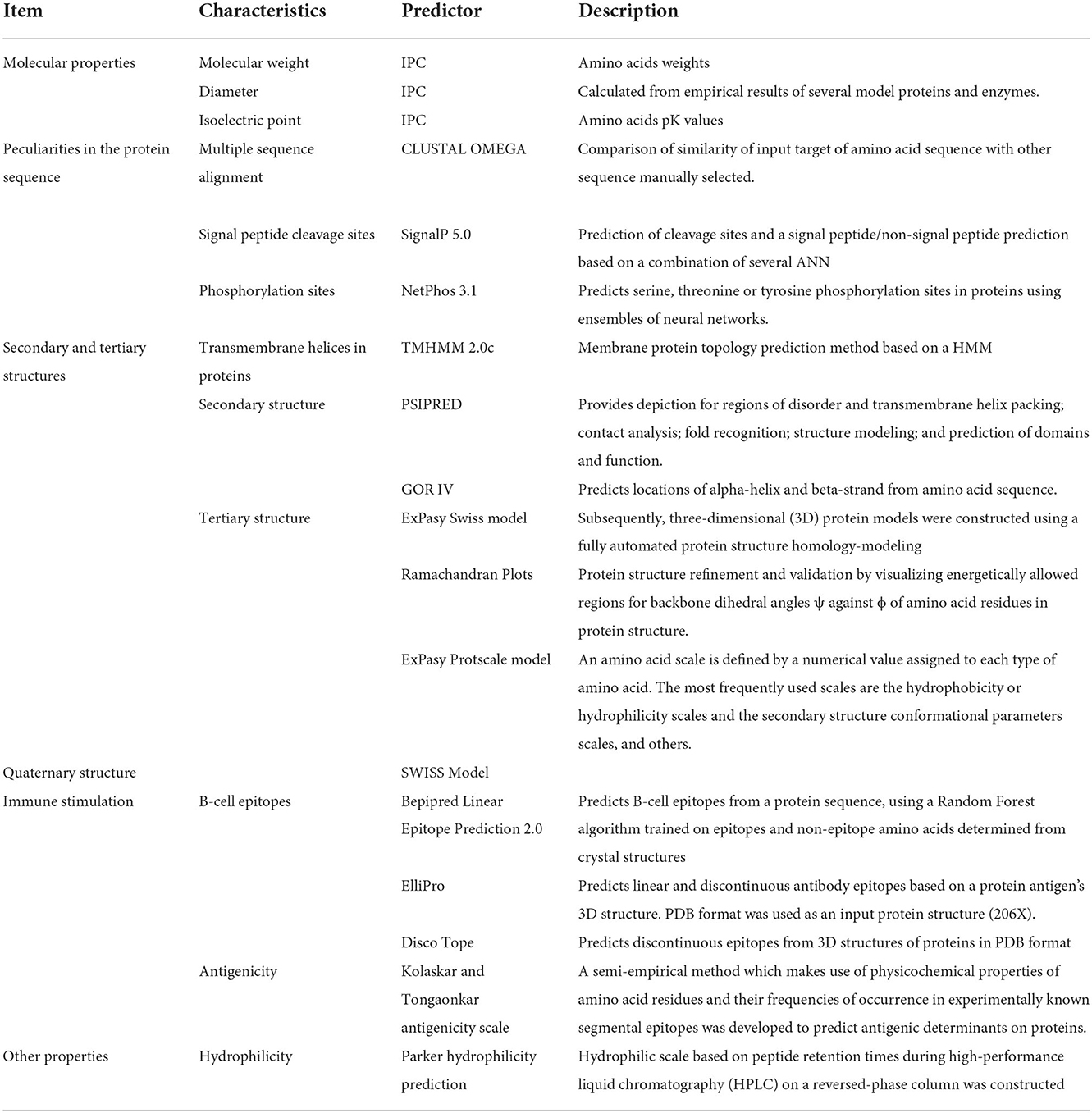
Methods: For this purpose, the secondary, tertiary, and quaternary structure of FhCatL1 protein were also predicted and analyzed using the SWISS-MODEL server. Validation of the modeled structures was performed by Ramachandran plots. The antigenic epitopes of the protein were predicted by IEDB server.
Results and discussion: Our findings revealed the low similarity of FhCatL1 with mammalian CatL1, lacking signal peptides or transmembrane domain, and the presence of 33 phosphorylation sites. Also, the containment of FhCatL1 for many topological, physico-chemical, immunological properties that favored its function of solubility and interaction with the immune components were reported. In addition, the earlier worldwide reports documented the high efficacy of FhCatL1 as a diagnostic and vaccine antigen in different animals. Altogether, FhCatL1 is considered an excellent candidate for using in commercialized diagnostic assays or vaccine products against fascioliasis in different animal species. Our assessment also included FgCatL1 and reported very similar findings and outputs to those of FhCatL1.
Introduction
The liver flukes Fasciola hepatica and F. gigantia are common foodborne trematode parasites widespread worldwide. They affect a wide variety of host species causing fascioliasis. Fasciola parasites are major pathogens of ruminants and have a substantial impact on the health and wellbeing of animals. Severe infections can be fatal as a result of widespread liver damage caused by the migration of large numbers of young flukes. Chronic infections lead to anemia, weight loss and poor condition. However, low-grade infections are common and asymptomatic and are associated with slower growth and lower milk production (1, 2). People or animals become infected by Fasciola parasites via the ingestion of food contaminated with encysted metacercariae on aquatic plants or vegetables. According to the World Health Organization, several millions of people are at risk of fascioliasis and 2.4 million people in more than 70 countries are infected with this parasite (3).
Accurate diagnosis and vaccine development against virulent Fasciola spp. are a critical part of the control program of fascioliasis particularly with an increase in prevalence of liver flukes infection in human and animals. This elevation is predicted to aggravate because of climatic changes and emergence of triclabendazole-resistant Fasciola (4, 5). Usually, fascioliasis is diagnosed by coprology, e.g., sedimentation techniques. Coprology is easy to perform, but has a generally low sensitivity (30–70%), especially in cattle, and can only detect patent infections (6). Indirect enzyme-linked immunosorbent assay (ELISA) is another test for detection of Fasciola infections via screening for specific antibodies in serum and milk samples. Traditionally, these ELISAs utilize the crude fluke excretory/secretory (ES) proteins that possess strong antigenic characteristics. These tests demonstrated a high sensitivity (86.1–100 %) and specificity (70–99.3 %) (7). Among ES antigens, proteases are encountered in 73 % of the total protein content comprising cathepsin L, cathepsin B, serinoprotenases, metalloproteases, and legumains (8).
Cathepsin L1 (CL1) and CL2 are the most abundant of proteases in ES antigen and the two proteins accounting for 67 % and 28% of total cathepsin L proteins, respectively (9). However, CL1 has been the antigen of choice for diagnostic tests and as a potential vaccine candidate in numerous animal models (10, 11). In a more recent study, proteomic analyses revealed that the recombinant and native cathepsin L zymogens contain conserved, highly antigenic epitopes that are conformationally dependent. Additionally, the diagnostic capacities of cathepsin L zymogens were confirmed using serum and fecal panel of samples, suggesting an excellent efficacy as markers of infection and for monitoring treatment efficacy (12).
Bioinformatics is one of the most current study fields to better address and monitor biological challenges (13). It is frequently used to evaluate the expression of genes and proteins as well as to forecast their general properties such as antigenicity and immunogenicity. These factors will significantly increase our understanding of these molecules and help scientists choose the best proteins or particular epitopes for studies on vaccines or medical diagnosis.
Epitopes are typically identified by showing that certain peptide fragments and certain proteins have antigenic reactivity. Since the accessibility, hydrophilicity, and mobility of polypeptide chain segments have been linked to the position of epitopes in proteins, it is conceivable to infer from the primary structure, in which linear peptides are most likely to correspond to those epitopes (14). In order to gain some insights into the molecular structure of its insoluble counterpart, secondary structure and antigenicity predictive methods have been applied to the amino acid sequences of different origins (15). This indicates that such a mechanism can be applied for diagnostic or immunogenic applications, such as our topic. It has been tried numerous times to infer the location of antigenic sites in proteins from specific traits of their primary, secondary, or tertiary structures. According to recent findings, the antigenicity definition varies depending on the type of probe employed to analyze it (16). Without knowledge of the protein's tertiary structure or the presence of discontinuous epitopes, the specificity of an immune response might also be accurately predicted for vast protein sections. Instead, antigenicity is a local attribute of the protein sequences, and the immune response's specificity and antigenicity are determined by the protein sequences' composition, secondary structure, solvent accessibility, and evolutionary conservation (17). Due to its continuous epitope distribution, we propose that the tertiary protein structure may have the same antigenicity as the primary structure. A linear peptide fragment of the epitope is highly cross-reactive with the matching antibodies. These epitopes are single-stretch polypeptide chains that are referred to as linear or continuous epitopes (18). By applying additional features, the B-cell epitope prediction task can be further enhanced. For instance, understanding the secondary structure and solvent accessibility of proteins in their three-dimensional (3D) form is essential. All of the evidence points of the efficient antigenicity being shared by primary and tertiary protein structures.
Few studies have analyzed the potentials of FhCatL1 as diagnostic or vaccine antigens using bioinformatic tools and much less for FgCatL1 (19, 20). Nevertheless, such studies have only focused on certain properties and using few analysis tools. Thus, the present study is providing inclusive and exclusive information on the physico-chemical, antigenic and immunogenic properties of F. hepatica cathepsin L1 (FhCatL1) protein using multiple bioinformatic analysis tools and several online web servers. In addition, data of bioinformatic analyses have been compared with the previous heritage of available studies investigated the potentials of FhCatL1and FgCatL1in immnunodiagnostic and vaccine researches.
Materials and methods
Amino acid sequence retrieval
The amino acid sequence for F. hepatica cathepsin L1 (FhCatL1) was obtained in FASTA format from the GenBank (AJ279092.1) database at https://www.ncbi.nlm.nih.gov. Then, using the corresponding cathepsin L1 proteins from FhCatL1, as well as those from F. gigantica (JQ342985.1), humans (BC142983.1), and cattle, multiple sequence alignment was performed using Clustal Omega (https://www.ebi.ac.uk/Tools/msa/clustalo/) (BC134741.1). For full matching with FhCatL1, first 15 amino acid (MRLFILAVLTVGVLG) was deleted from our analyzed F. gigantica (JQ342985.1).
Post-translational modification (PTM) sites and transmembrane domains
To identify the signal peptide in the sequence, the SignalP online tool (https://services.healthtech.dtu.dk/service.php?SignalP-5.0) was utilized. A Sec signal peptide (Sec/SPI), a Lipoprotein signal peptide (Sec/SPII), a Tat signal peptide (Tat/SPI), or no signal peptide at all can be found in the protein (Other). Additionally, using the TMHMM 2.0 online server (https://services.healthtech.dtu.dk/service.php?TMHMM-2.0), putative transmembrane domains of FhCatL1 and FgCatL1 were predicted (21). Using the online isoelectric point calculator at http://isoelectric.org, the isoelectric point (pI) and molecular weight of FhCatL1 and FgCatL1 was calculated. The molecular weight and diameter of the selected amino acids from FhCatL1 and FgCatL1of number 311 were determined.
Physical, chemical and morphometric characteristics
The NetPhos 3.1 server (https://services.healthtech.dtu.dk/service.php?NetPhos-3.1) used ensembles of neural networks to show the locations of phosphorylation sites for serine, threonine, and tyrosine. Two web servers were used to clarify the FhCatL1 and FgCatL1 secondary structure, including the Garnier-Osguthorpe-Robson (GOR IV) servers (https://npsa-prabi.ibcp.fr/cgi-bin/npsa_automat.pl?page=/NPSA/npsa_gor4.html) and position specific iterated prediction (PSIPRED) (22, 23), based on PSI-BLAST results (http://bioinf.cs.ucl.ac.uk/psipred). Then, three-dimensional (3D) protein models were created using SWISS-MODEL, a fully automated online server for protein structure homology modeling that may be found at (https://swissmodel.expasy.org/) (24). Using the Ramachandran plot analysis used by the Swiss model, the tertiary structure was further refined and validated for both FhCatL1 and FgCatL1. This was done based on the percentage of residues in the outlier and preferred regions. Additionally, using the ProtScale tool (https://web.expasy.org/protscale/), graphical representations of linear epitopes were created based on average flexibility, beta-turn, alpha-helix, hydrophobicity, and the percentage of accessible residues and molecular weight (25).
Prediction of B-cell epitopes, antigenicity, and hydrophilicity
Based on the physicochemical characteristics of amino acids including hydrophilicity (26), and antigenicity (27), the Immune Epitope Database (IEDB) (http://tools.immuneepitope.org/bcell/) was employed by the IEDB server to predict the B-cell epitopes. ElliPro predictor was used to analyze linear and conformational (discontinuous) epitopes based on 3D structures prediction of FhCatL1. Crystal Structure of ProCathepsin L1 from Fasciola hepatica (206X) that showed high similarity with our target sequence of FhCatL1, as it is available as PDB file, the required input data for ElliPro searches was used for further analysis of FhCatL1 (http://tools.iedb.org/ellipro/). Similarly, Disco Tope tool that can predict the conformational antibody epitope via 3D structures was used employing the same the protein model 206X (http://tools.iedb.org/discotope/). A summary for the predictor servers used in the current study for analyses of different properties of FhCatL1 was illustrated in Table 1.
Results and discussion
Multiple sequence alignment and molecular weight
The amino acid sequence of FhCatL1 protein was obtained from GenBank accession ID: AJ279092.1which was submitted by Cornelissen et al. (28). Then, CLUSTALO-based multiple sequence alignment of FhCatL1 was applied against cathepsin L1 of F. gigantica, as well as human and cattle amino acids as examples for highly susceptible host species for fascioliasis. Similarity was 94.2, 44.4, and 40.2% for cathepsin L1 of F. gigantica, human and cattle, respectively (Supplementary Figures S1A,B). These results demonstrated the high similarity of FhCatL1 with other Fasciola cathepsin L proteins and high differences with those of human or cattle suggesting the low probability of cross reaction with samples of such animal hosts.
Signal peptide, transmembrane domains, and isoelectric point
Based on the SignalP-5.0 and TMHMM output, no signal peptides or transmembrane domain were found for FhCatL1 (Supplementary Figures S2A–D). Our results based on transmembrane domain analysis suggest the cytoplasmic localization of FhCatL1 and FgCatL1. Cytosolic proteins include large complexes of enzymes that are responsible for several metabolic pathways, protein biosynthesis, and other important cell-signaling processes. Indeed, nuclear or cytoplasmic protein extracts are important for various applications, including RNA binding, in vitro transcription, mRNA splicing, or gene-expression studies. Thus, the antibody targeted cytosolic protein can serve as a molecular tool to enhance our knowledge of protein–protein interactions between Fasciola parasites and the host cell machinery (29). The theoretical average of pI of FhCatL1 was 5.2. Using all predictors, the pI was not lower than 5 except for Patrickios; pI = 3.248. In addition, the theoretical average of pI of FgCatL1 was 5.11 and Patrickios pI was 2.93 (Supplementary Figures S3A,B). The amino acids sequences of FhCatL1 protein include 311 amino acid residues with an estimated molecular weight of 35.2 kDa (antigens which have MW of < 5–10 KDa are considered as weak immunogens) (30), with a diameter equal to 4.67487 nanometer (nm). Similarly, FgCatL1 (JQ342985.1) consists of 311 amino acids with estimated MW of 35.2 kDa with similar diameter to FhCatL1 (Supplementary Figures S3C,D).
Two major components are affecting the production level of the recombinant protein; the presence of signal peptides and secretory machinery. The signal sequence is often a short peptide that carries information about the protein target and secretion pathway zip codes at the N-terminus of the most recently made proteins. Additionally, the antigen presentation would be improved by the absence of any potential transmembrane domains in order to stimulate humoral and cell-mediated immunity as well as a quick response (21, 31). A number of analytical biochemistry and proteomics procedures, such as 2-D polyacrylamide gel electrophoresis or capillary isoelectric focusing used in conjunction with high-throughput mass spectrometry, depend on the accurate estimate of the isoelectric point (pI) through the amino acid sequence. Additionally, protein crystallization experiments can benefit from pI estimation (32).
Physico-chemical and morphometric characteristics
NetPhos server was used for detection of phosphorylation sites in FhCatL1 protein based on serine, threonine, and tyrosine amino acids. Analysis results revealed that there were 33 phosphorylation sites in the FhCatL1 protein sequence, with 16 serine (S), six threonine (T), and eleven tyrosine (Y) as shown in Figures 1A,B. Consistently, analysis of FgCatL1 revealed 33 phosphorylation sites with 16 serine (S), 3 threonine (T), and 14 tyrosine (Y) (Supplementary Figure S4). The existence of phosphorylation sites indicates the high liability of protein for post translational modifications (PTMs) processes. PTMs in eukaryotic cells, such as the cells of parasites, are significant for choosing the correct expression system in the development of recombinant proteins and might affect the biological function of the protein (33, 34).
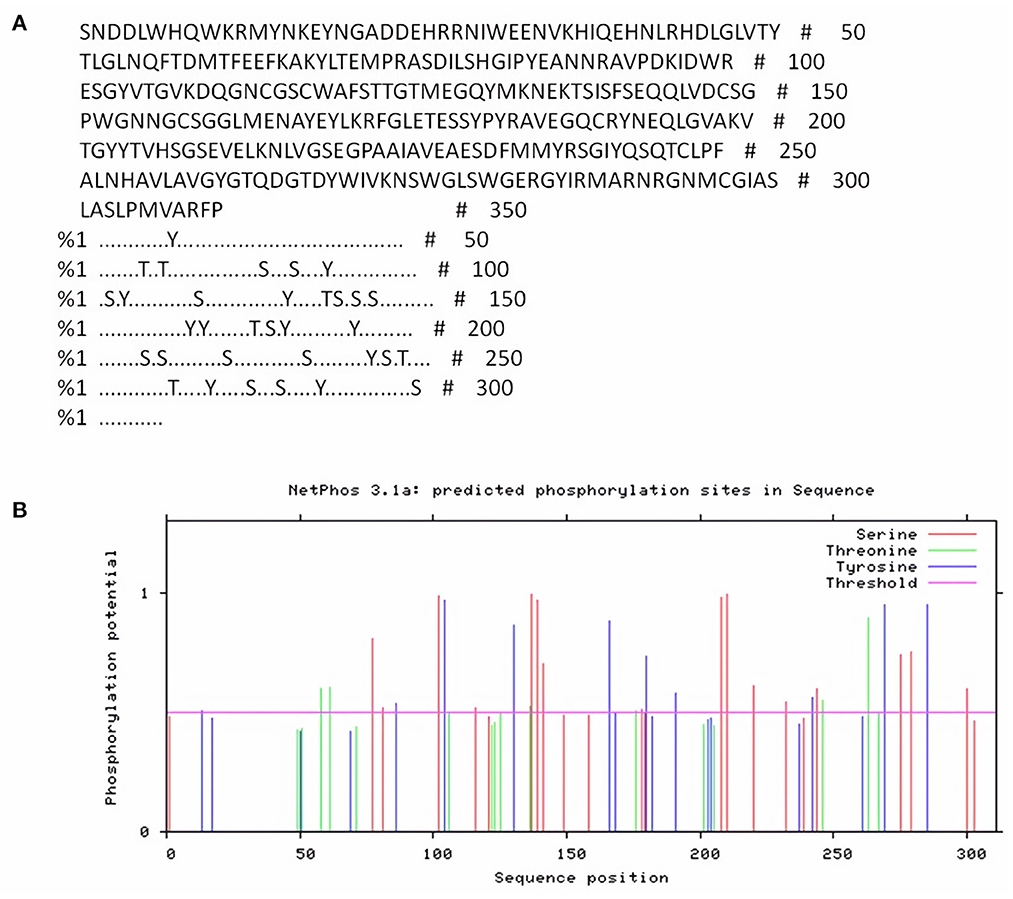
Figure 1. NetPhos server output for FhCatL1 phosphorylation sites. (A) The number of predicted sites, based on S (serine), T (threonine) and Y (tyrosine); (B) Prediction diagram of FhCatL1 phosphorylation sites (https://services.healthtech.dtu.dk/service.php?NetPhos-3.1).
The FhCatL1 protein contained three main secondary structure components, consisting of 53.38 % random coil, 24.44 % extended strand, and 22.19 % alpha helix, as shown by PSIPRED and GOR IV servers. For FgCatL1 amino acid sequences encompasses 52.41 % random coil, 25.08 % extended strand, and 22.51 % α-helix in secondary structure (Supplementary Figures S5, S6). Proteins' primary biological behavior is primarily determined by their spatial organization. The sequence of hydrogen bonding between carboxyl oxygen and amino hydrogen atoms in a polypeptide chain, with α-helices and 2D-structures being the most frequent types, typically determines the secondary structure (35).
Moreover, three-dimensional (3D) protein models were constructed for FhCatL1and FgCatL1using a fully automated protein structure homology-modeling web server called SWISS-MODEL. In total, 50 SWISS-MODEL templates were made for FhCatL1 protein sequence, among which a single model highly matching our target was built. The model showed highest coverage and 92.26% sequence identity. Global model evaluation GMQE and QMEANDisCo of any analyzed model gives an overall model quality measurement between 0 and 1, with higher numbers indicating higher expected quality. GMQE and QMEANDisCo in our model was 0.96 and 0.93, respectively, suggesting high quality of expected model measurement and assessment (Figures 2A–D). The significance of the QMEAN Z-score as an indicator of protein stability was further highlighted by the fact that our model had significantly high QMEAN Z-scores in a pairwise comparison with their homologous mesophilic counterparts. In our case, QMEAN Z-score was −1.12 which is close to zero suggesting high accuracy and reliability of protein modeling (Figure 2E). In addition, our target model was distributed in the same area when compared with non-redundant set of protein data base reference structures (Figure 2F). Red color in sequence alignment and amino acid sequence ANNRA, and low indentation at low quality estimate curve near 0.4 are showing area of instability which might be prone to mutation. Three dimensional structures was also applied for FgCatL1 and found similar data and findings to FgCatL1. Crystal Structure of ProCathepsin L1 from Fasciola hepatica was found and used as a model for further template to FgCatL1 (Supplementary Figure S7).
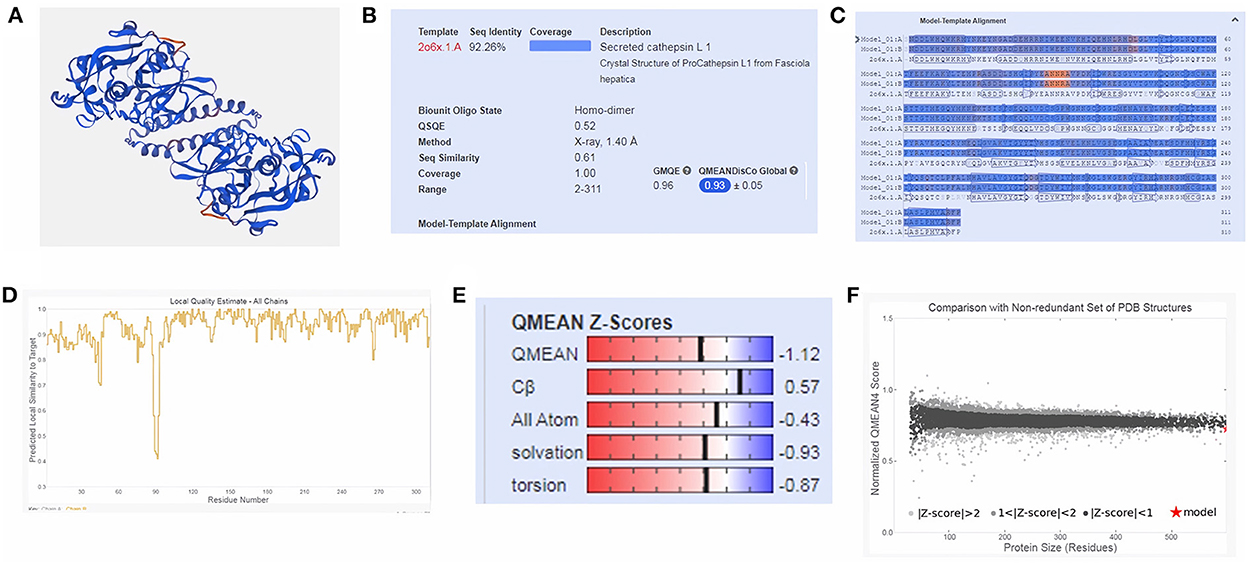
Figure 2. Tertiary structure of FhCatL1 using SWISS-MODEL server. SWISS-MODEL server results of (A) Computed three-dimensional model; (B) Sequence identity and coverage data; (C) Model-template alignment; (D) Local quality estimate; (E) Global quality estimate; (F) Comparison with non-redundant set of PDB structures (https://swissmodel.expasy.org/).
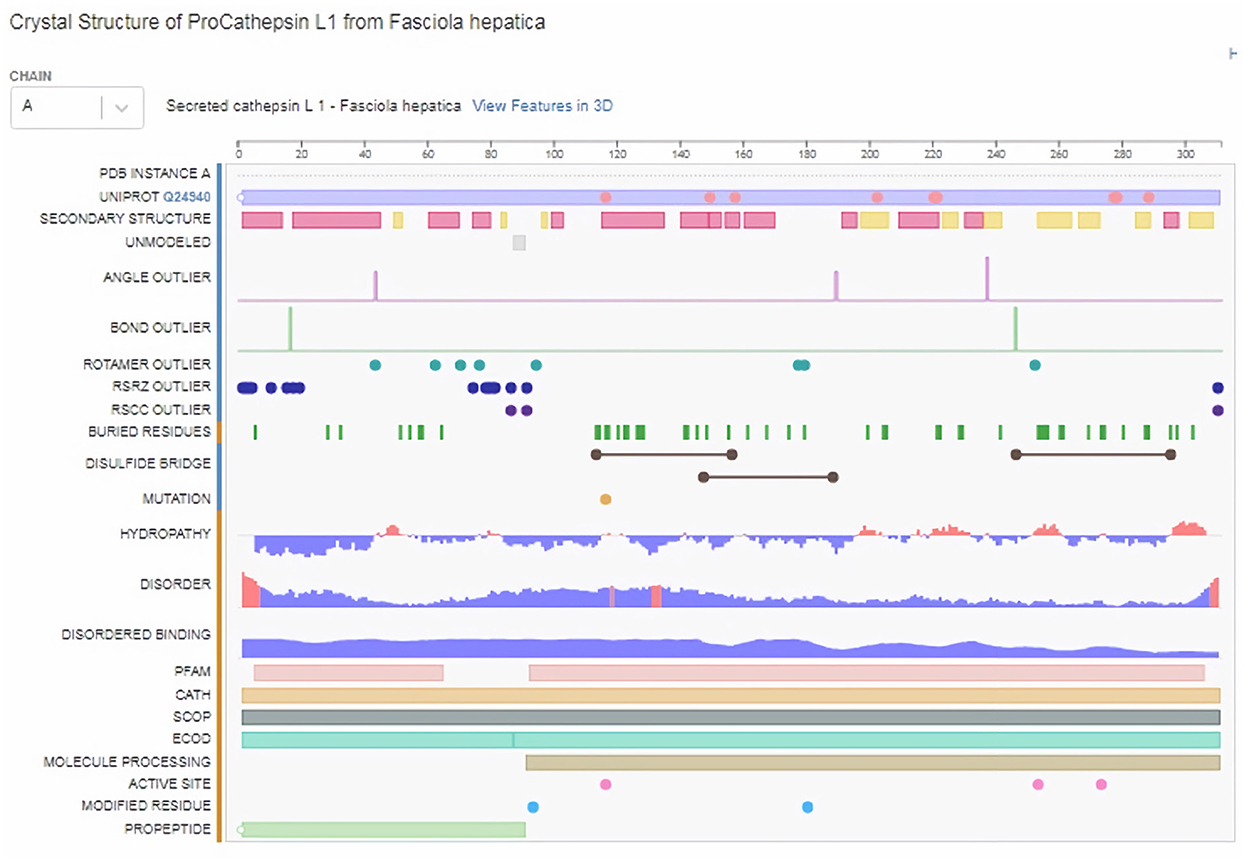
Subsequently, the chosen 3D model was analyzed via Ramachandran plot from SWISS-MODEL server. The model demonstrated MolProbity (0.92), Clash score (0.21), Poor rotamers (1.0) and Ramachandran favored (95.78%), no Ramachandran outliers, C-Beta deviations or bad bonds, Rotamer outliers (1.16%), bad angels (30 / 6856) and twisted proline (2/18) (Figure 3). These results indicate the high quality of 3D model of FhCatL1. Also, Ramachandran plot was applied for FgCatL1 revealed a very similar data to those obtained for FhCatL1 (Supplementary Figure S8). As such, to comprehend the influences between both structures and functions, assessment of 3D structure is the key aim of expecting a protein's nature (36, 37). More accurate mapping of different structure of amino acid residues in our 3D structure model was also analyzed (Figure 4). Furthermore, analysis of such model was conducted via quaternary structure either for degree of oligomerization, stoichiometry, topology, and the interface similarity (Figure 5). Our target sequence showed a homo-dimeric oligo state. High sequence similarity was observed when our target sequence of FhCatL1 was compared against conserved model Crystal Structure of ProCathepsin L1 from Fasciola hepatica (206X) at SWISS-MODEL repository for all tested characters of oligomerization, stoichiometry, topology, and the interface similarity based on quaternary structure analysis (Figures 5A–E). The dimeric forms of FhCatL1 have a stronger interface conservation signal with clade including 206X model with high similarity to our target sequence. This stronger conservation is observable using different evolutionary distance thresholds. Such knowledge about stoichiometry, topology, and the interface similarity aid in the understanding of FhCatL1 interactions with other biomolecules, and provide insights into their function in various biochemical processes, such as signal transduction, immune response or metabolism (38). As 206X sequence data is already publicly available at http://oligo.swissmodel.expasy.org, and at https://www.rcsb.org/sequence/2O6X, similar data can be obtained from these websites. However, our collection from other servers will assist the molecular biologists and biochemists by providing a detailed description and discussion for the physico-chemical properties of FhCatL1 protein. SWISS MODEL provides an automated tool for analysis of a protein sequence using different levels of structural forms particularly tertiary and quaternary structures (24). Further results regarding structural and physical properties were also obtained from protein data bank (PDB) after searching FhCatL1 protein (Figure 6). These results confirmed those of low values for outlier amino acid residues and additionally reported minimum liability for hydropathy, mutation and disorders. Recombinant proteins increasingly replace complete organisms in current immunotherapy and vaccination in order to stimulate therapeutic or protective immune responses. Protein structural stability is essential for the effective presentation of antigenic peptides on MHC, which is critical for inciting powerful immune responses (39). Thus, our data concerning the physical and chimerical properties, suggest the usefulness of FhCatL1 for further immunological investigations. A summary of physico-chemical and structural properties is shown in Supplementary Table S1.
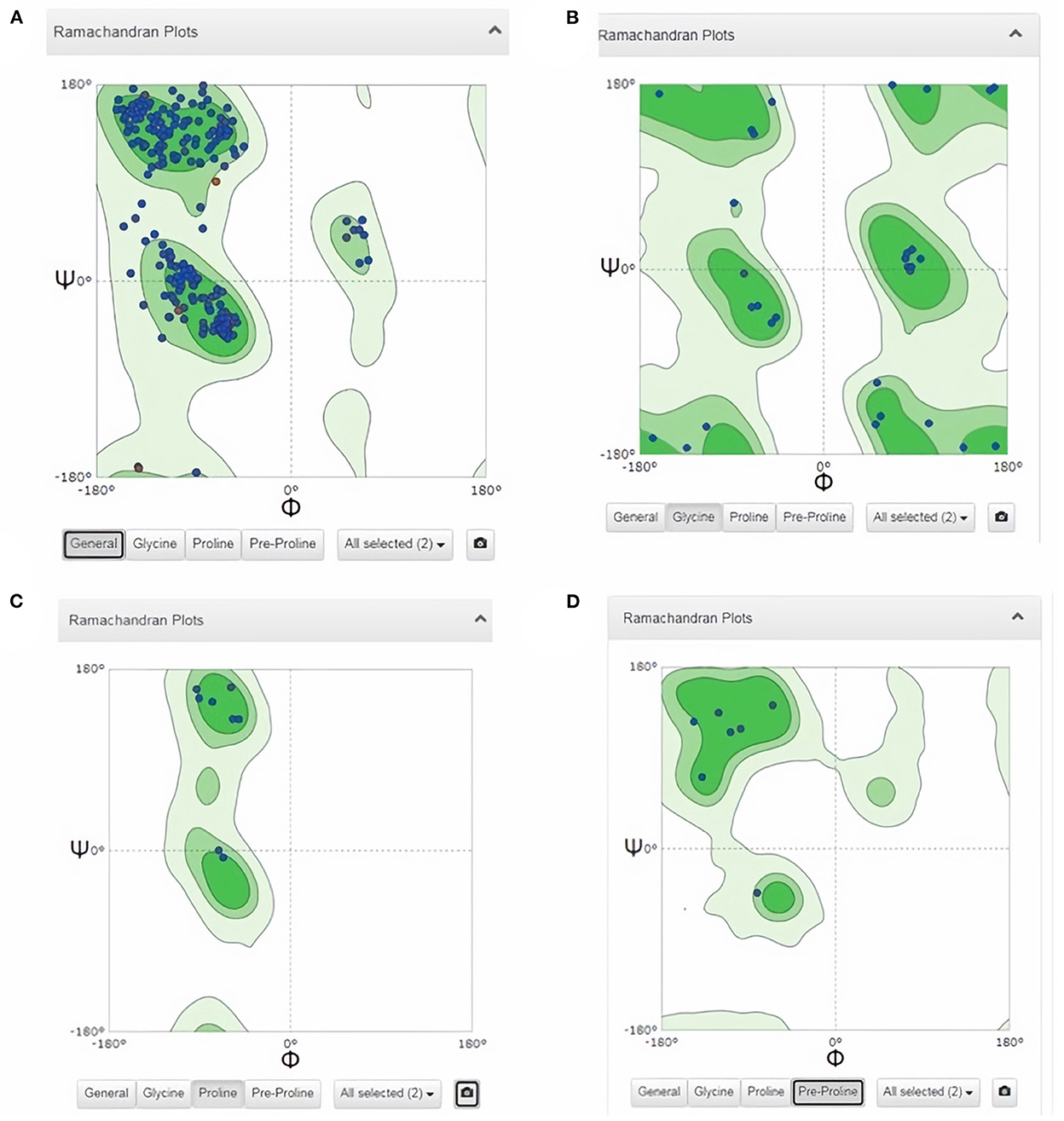
Figure 3. Analysis of 3D model of FhCatL1 via Ramachandran plot from SWISS-MODEL server. The model demonstrated MolProbity (0.92), Clash score (0.21), Poor rotamers (1.0) and Ramachandran favored (95.78%), no Ramachandran outliers, C-Beta deviations or bad bonds, Rotamer outliers (1.16%), bad angels (30/6856) and twisted proline (2/18). (A) General plot, (B) Glycine based plot, (C) Proline based plot, (D) PreProline based plot (https://swissmodel.expasy.org/).
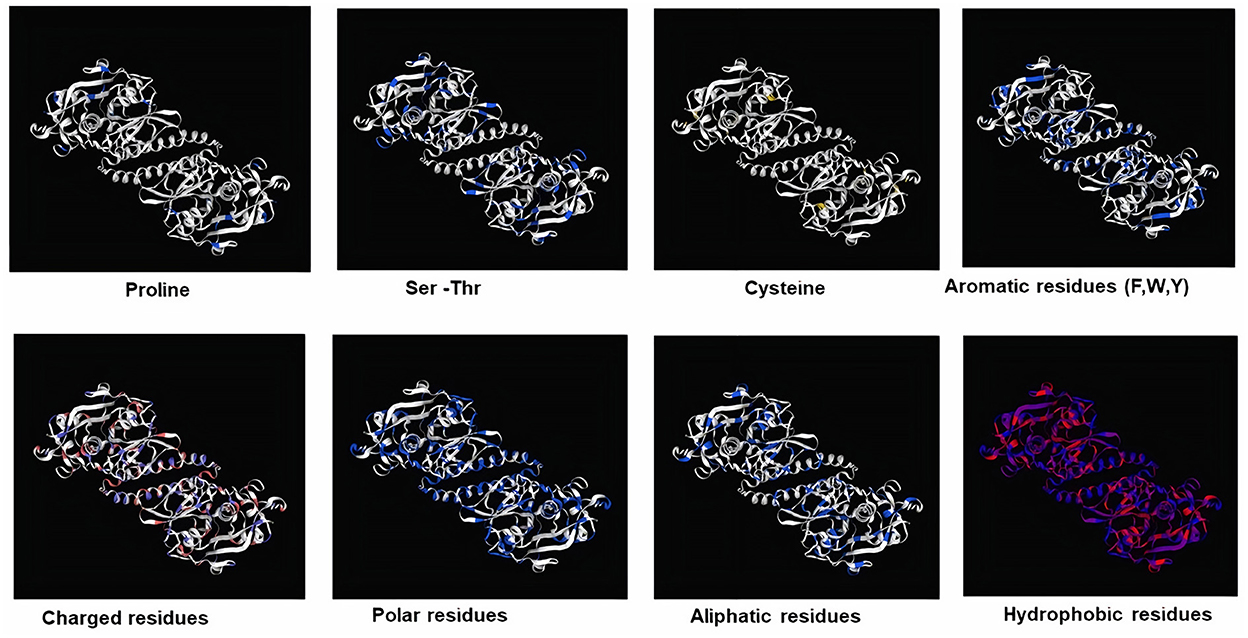
Figure 4. Mapping of different structure of amino acid residues in 3D structure model of FhCatL1 (https://swissmodel.expasy.org/).
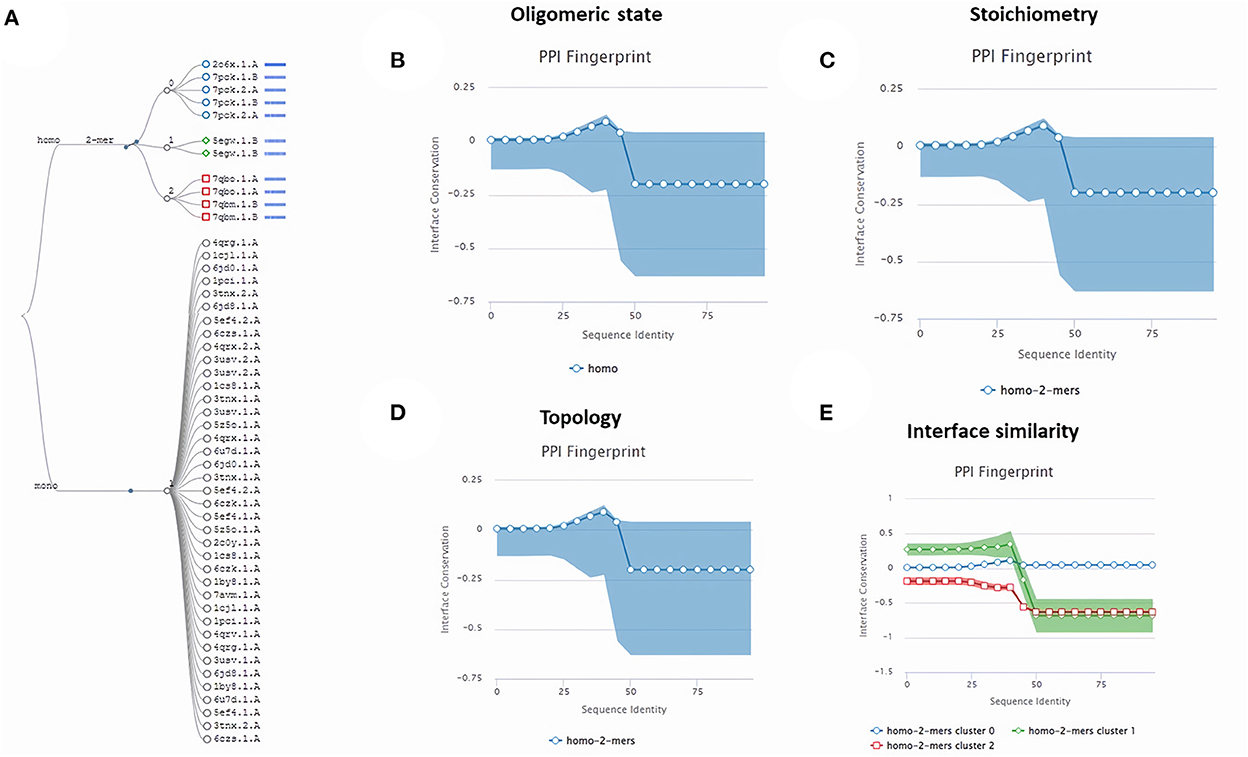
Figure 5. Quaternary structure analysis of FhCatL1 as a homo-dimer model (A) Structural clustering tree of FhCatL1 homologs with known structure. Each leaf is a template labeled with the PDB code and a bar indicating sequence identity and coverage (darker shades of blue refer to higher sequence identity). The decision tree follows the described levels of clustering: oligomeric state (B), stoichiometry (C), the topology of the complexes (D), and interface similarity (E). The PPI fingerprint curves of our oligo-dimeric target shows high consistency with PDB model code 206X with high consistency as shown oligomeric state, stoichiometry and topological characteristics. Also, our homo-dimer target showed a higher similarity for interface interactions with cluster 0 (blue circle) included 206X than cluster 1 (green rhomboid) and 2 (red square).
Immune-epitope analyses of FhCatL-1
ProtScale online tool was applied to predict linear B-cell epitopes for FhCatL1 and FgCatL1 according to hydrophobicity, alpha-helix, beta-turn, average flexibility, percent of accessible residue, and molecular weight (Supplementary Figure S10). Strong signals were obtained in all the plots of different scales by most amino acids residues suggesting the high presence of immune epitopes in our tested sequence of FhCatL1 and FgCatL1. High values estimated for epitopic locations with good hydrophilicity and potent antigenicity were found using the IEDB server. Bepipred Linear Epitope Prediction 2.0 predicted 11 anticipated peptides and a threshold of 0.5, with estimated lengths ranging from 3 to 55 amino acids for the linear B-cell epitope areas (Figure 7A). Kolaskar and Tongaonkar also predicted nine anticipated peptides with high antigenicity indices. Antigenicity with a threshold of 1.007, and an estimated amino acid range of 6 to 24 was observed (Figure 7B). Also, the hydrophilicity of FhCatL1 was detected by Parker Hydrophilicity Prediction scale showing high index of water solubility with threshold 1.877 (Figure 7C). Similar results were obtained when analyzing sequence of FgCatL1 using before mentioned predictors (Supplementary Figure S11). In addition, linear and discontinuous antibody epitopes based on a protein antigen's 3D structure was analyzed using ElliPro. This analysis server based on utilizing the PDB file of a certain protein not amino acids sequence as input data. Thus, the available protein file in PDB named 206X was used to assess the different epitopes of FhCatL1. As a primary analysis, ElliPro 2D chart was also illustrated which showed high prediction of epitope residues (Figure 7D). For epitope mapping, ElliPro 3D analysis tool was used to reveal the linear and conformational epitopes. For this purpose, chain A and B of the used PDB file of 206X.1A and 206.1B was assessed separately, respectively for more accurate prediction. ElliPro search revealed 9 predicted epitopes with showing 3D structure and Jmol visualization. The largest residues of the used protein model consist of linear epitopes with 21 to 49 amino acids with score ranged from 0.663 to 0.822 (Figure 8). Regarding the conformational epitopes, ElliPro analysis demonstrated 3 amino acid residues with score ranged from 0.64 to 0.751 (Figure 9). Disco Tope is another analysis tool used to predict the conformational antibody epitope based on 3D structures prediction and using PDB file as input data. Crystal Structure of ProCathepsin L1 from Fasciola hepatica (206X.1A or 206X.1B) was assessed followed by visualization of 2D charts and Jmol visualization of the predicted epitopes. As shown in Figure 10, 2D chart and Jmol display the structure with high positive predictions (74 residues from total 310) highlighted in yellow in both panels. These results revealed the high potentials of FhCatL1 for interacting with immune cells and generating specific antibodies.
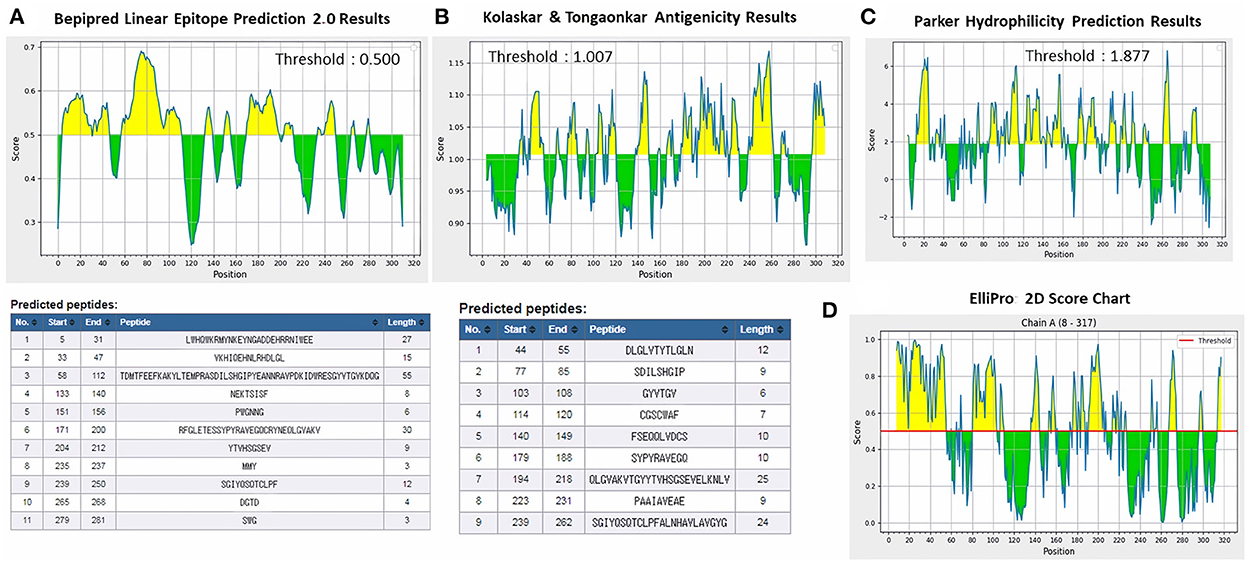
Figure 7. Immunogenicity, antigenicity, and hydrophilicity of FhCatL1 protein. (A) Bepipred linear epitopes prediction; (B) Antigenicity; (C) Hydrophilicity; (D) ElliPro: 2D Score Chart epitope prediction. x-axis and y-axis represent position and score, respectively. The horizontal line indicates the threshold or the average score. Yellow colors (above the threshold) indicate favorable regions related to the properties of interest. Green color (below the threshold) indicates the unfavorable regions related to the properties of interest. The results were analyzed by the Immune Epitope Database (IEDB) (http://tools.immuneepitope.org/bcell/).
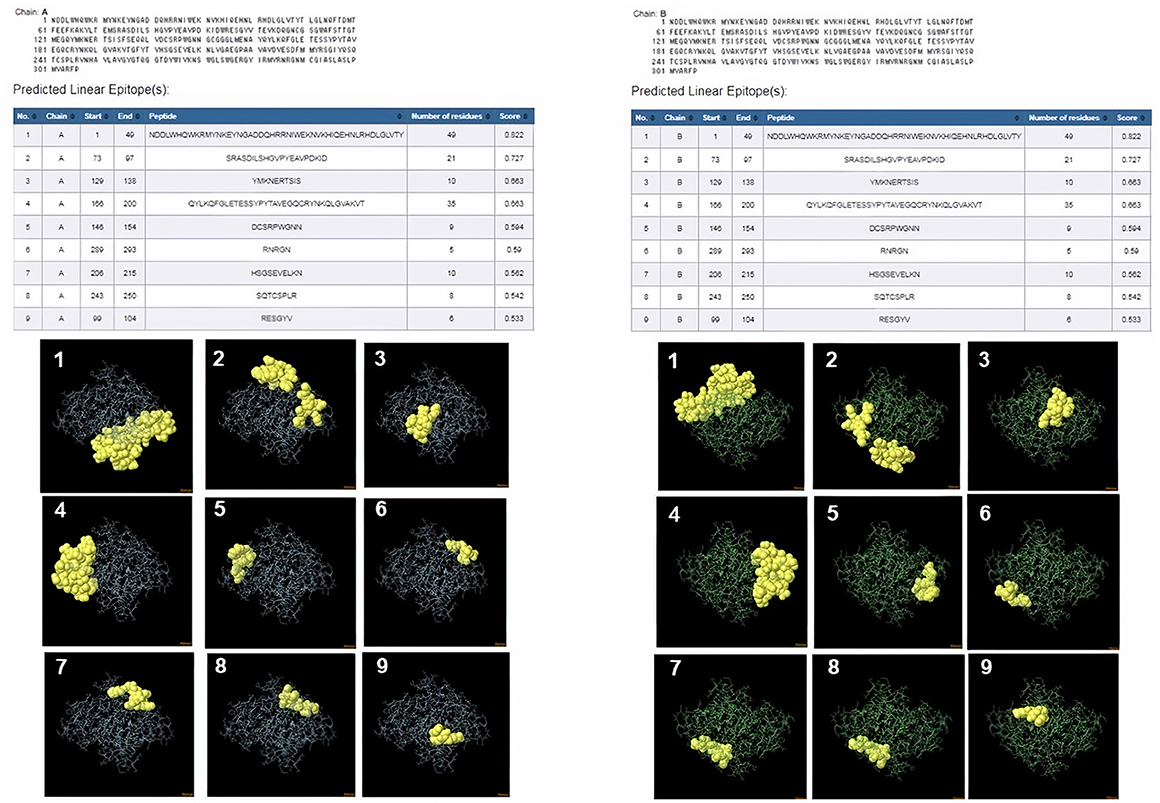
Figure 8. ElliPro: Linear antibody epitope 3D structures prediction of FhCatL1. Crystal Structure of ProCathepsin L1 from Fasciola hepatica (206X.1A) chain A PDB file was used as input data for ElliPro searches. Input sequence, table of predicted epitopes, and Jmol visualization of the 9 predicted epitopes shows the epitope residues are in yellow and the rest of the protein is in violet (left panel). Crystal Structure of ProCathepsin L1 from Fasciola hepatica (206X.1B) chain B PDB file was used as input data for ElliPro searches. Input sequence, table of predicted epitopes, and Jmol visualization of the 9 predicted epitopes shows the epitope residues are in yellow and the rest of the protein is in green (right panel) (http://tools.iedb.org/ellipro/).
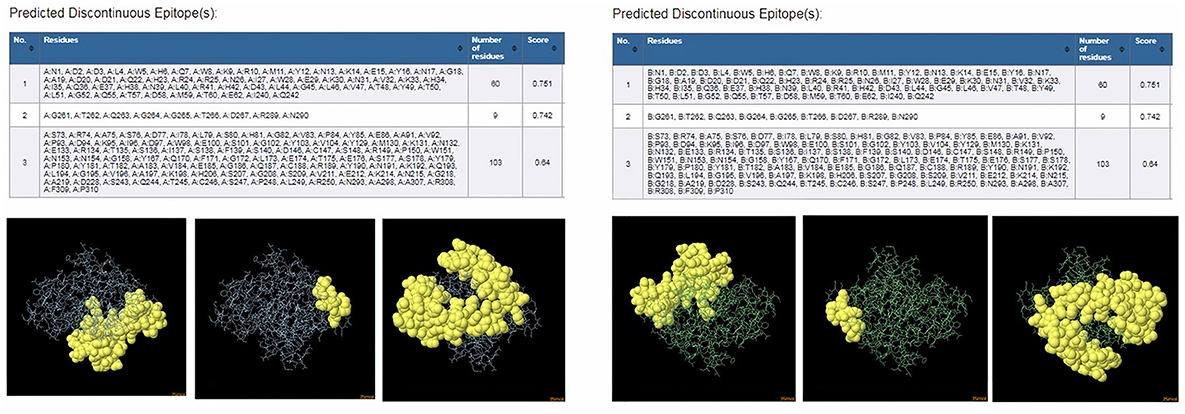
Figure 9. ElliPro: Discontinuous (conformational) antibody epitope 3D structures prediction of FhCatL1. Crystal Structure of ProCathepsin L1 from Fasciola hepatica (206X.1A) chain A PDB file was used as input data for ElliPro searches. Table of predicted epitopes, and Jmol visualization of the 3 predicted epitopes shows the epitope residues are in yellow and the rest of the protein is in violet (left panel). Crystal Structure of ProCathepsin L1 from Fasciola hepatica (206X.1B) chain B PDB file was used as input data for ElliPro searches. Table of predicted epitopes, and Jmol visualization of the 3 predicted epitopes shows the epitope residues are in yellow and the rest of the protein is in green (right panel) (http://tools.iedb.org/ellipro/).
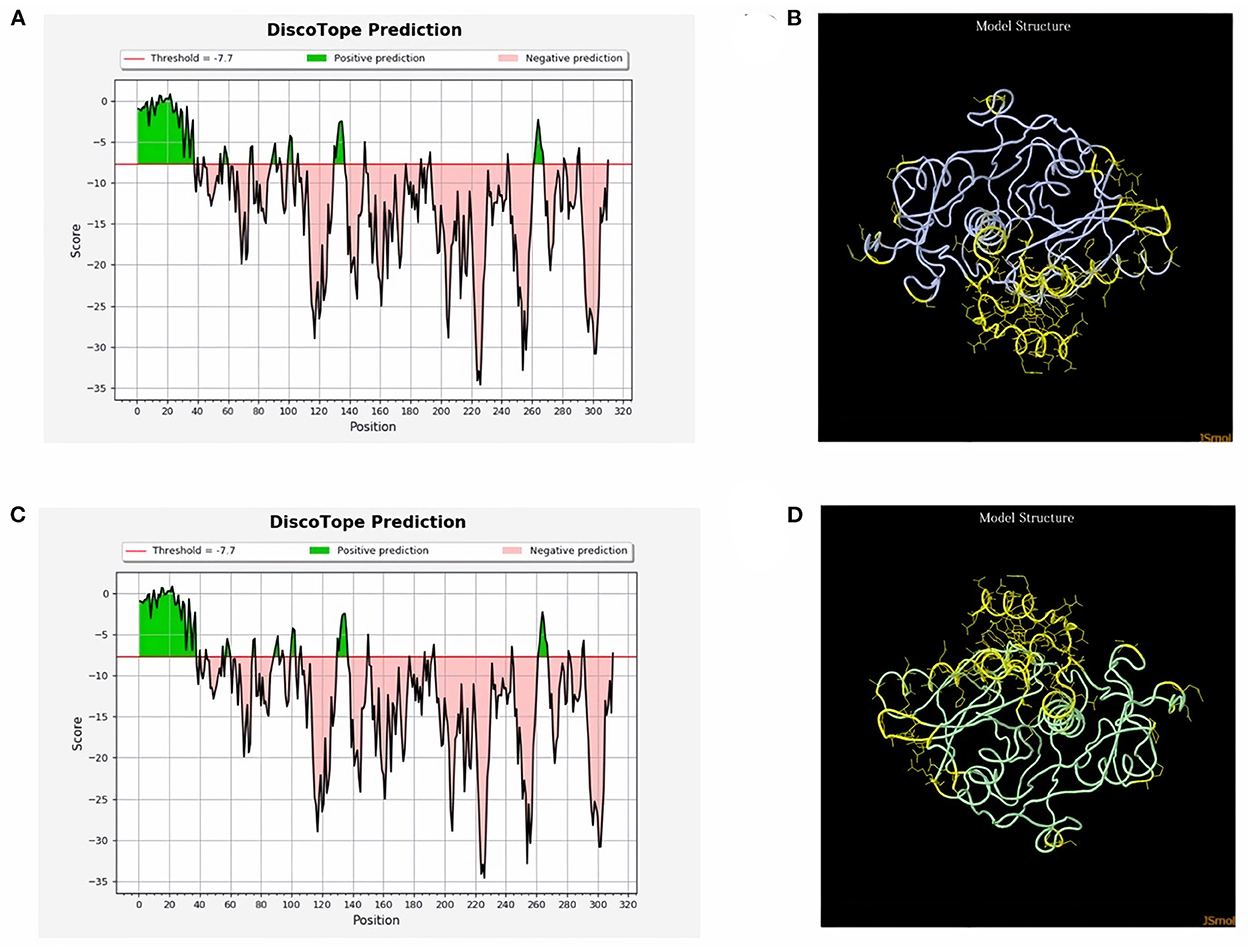
Figure 10. Disco Tope: Discontinuous (conformational) antibody epitope 3D structures prediction of FhCatL1. Crystal Structure of ProCathepsin L1 from Fasciola hepatica (206X). Chain A PDB file was used as input data for Disco Tope searches (https://www.rcsb.org/structure/2O6X) for visualization of 2 D chart (A) and Jmol visualization (B) of the predicted epitope. Chain B PDB file was used also as input data for visualization of 2 D chart (C) and Jmol visualization (D) of the predicted epitope shows the epitope residues are in yellow and the rest of the protein is in violet (http://tools.iedb.org/discotope/).
The interaction between antibodies and antigens is one of the most important immune system mechanisms for clearing infectious organisms from the host. Antibodies bind to antigens at sites referred to as B-cell epitopes. Identification of the exact location of B-cell epitopes is essential in several biomedical applications such as; rational vaccine design, development of disease diagnostics and immunotherapeutics. However, experimental mapping of epitopes is resource intensive making in silico methods an appealing complementary approach (40). An epitope for a protein antigen may be a brief peptide from the protein sequence, known as a linear epitope, or it may be a patch of atoms on the surface of the protein, known as a discontinuous or conformational epitope. The goal of discontinuous epitope prediction is to create a molecule that can mimic the structure and immunogenic properties of an epitope and replace it either in the process of antibody production—in this case, an epitope mimic can be thought of as a prophylactic or therapeutic vaccine—or antibody detection in medical diagnostics or experimental research. While, linear epitopes can be directly used for the design of vaccines and immunodiagnostics (41, 42).
Herein, we provided essential information on the physico-chemical, antigenic, and immunogenic capacities of FhCatL1 and much less extent of FgCatL1. This analysis was variably relying on the availability of data in three major data set repositories including NCBI, SWISS-MODEL, and PDB. However, the data presented here is providing numerous beneficial insights on the characterization of FgCatL1 from different aspects ranged from structural to advanced antigenic or immunogenic properties. Nevertheless, these data have the limitations of generation using only bioinformatic or computed analysis. Thus, in the following section, a literature review was conducted to correlate the obtained bioinformatic findings of FhCatL1 or FgCatL1 with those of realistic works conducted in the immunodiagnostics and preventive studies.
Potentials of Fasciola spp. cathepsin L1 as a diagnostic and vaccine antigen based on previous reports
In addition to zoonotic importance, fascioliasis of livestock has a direct major financial impact globally through production losses. Such losses can include poor weight gain or productivity, condemnation of liver, lower commercial return, and inability to trade nationally and internationally (1, 2).
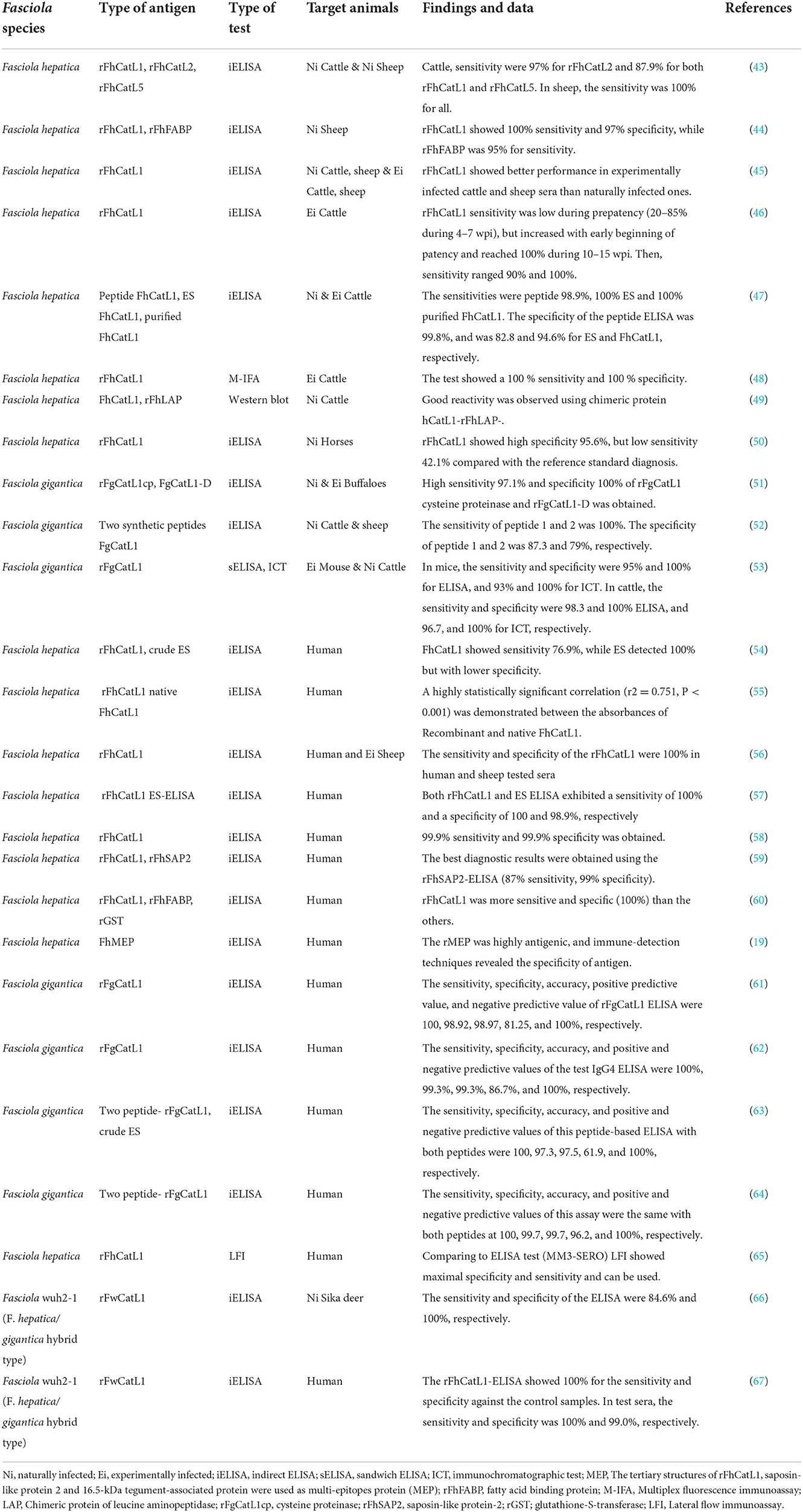
The accurate diagnosis of Fasciola infections is central to studying the epidemiology of fascioliasis and the surveillance and control of this disease. Two main species of Fasciola named F. hepatica and F. gigantica have been studied and showed great impact on human and animal health. Numerous studies have reported the aspects of the evaluation and validation of CatL1 antigen in diagnosis of different types of fascioliasis in various animals. In case of F. hepatica, recombinant CatL1 antigen (rFhCatL1)-based indirect ELISAs demonstrated high efficacy in detection of specific antibodies in infected animals including sheep (43–45). In addition, this method (rFhCatL1-based ELISA) showed excellent performance against sera of naturally or experimentally infected cattle (43, 45, 46). Also, Cornelissen et al. (47), compared indirect ELISAs of peptide and purified FhCatL1 against excreted secreted (ES) antigens of F. hepatica in naturally and experimentally infected cattle. This study showed a sensitivity of 98.9% for the peptide, 100% for purified FhCatL1 and 100% for ES. For specificity, values of 99.8% were found for the peptide, 94.6% for purified FhCatL1, and 82.8% for the ES antigens. Noteworthy, this study used a residue containing 15 amino acids, was predicted in our study as a sequence included in our detected epitope number 3 ranged from 58-112 of FhCatL1 used sequence. In such study, selected peptide sequence was based on immunoproteomics not on bioinformatics as in our case. An immunogenic antigen released in vitro by F. hepatica was purified. After purification the sequence of the first 20 N-terminal aa of this protein showed considerable homology with cathepsin L-like proteinase Cornelissen et al. (47). Good reactivity was observed also against cattle sera using multiplex fluorescence immunoassay (MFI) based on rFhCatL1 (48), and western blotting relying on chimeric protein of leucine aminopeptidase (LAP) and FhCatL1 (49). In the later study, FhCatL1 (GenBank CAC12806.1) that is matching and identical to our used protein (GenBank AJ279092.1) was tested, and the constructed chimeric protein included amino acid sequence from 173 to 309 which includes 6 of our identified epitopes consisting of 61 amino acids.
Moreover, rFhCatL1- ELISA was tested against naturally infected horse sera and showed high specificity of 95.6%, but low sensitivity of 42.1% compared with the reference standard diagnosis (50). Not only FhCatL1 but also F. gigantica cathepsin L1 (FgCatL1) antigen has proven its potential in discriminating between infected and non-infected sera collected from susceptible animals. In this regard, various forms of FgCatL1 could detect fascioliasis in buffaloes, sheep, or cattle sera either using rFgCatL1 (51), synthetic peptide (52), or monoclonal antibody against rFgCatL1 (53) based ELISAs, respectively.
Even for human sera, FhCatL1 and FgCatL1 have demonstrated an excellent ability in detecting specific antibody in infected patients with very low cross-reactivity. Many studies have utilized the indirect ELISAs based on rFhCatL1 (19, 54–60). Consistently, FgCatL1 based ELISAs showed an optimal performance against human sera when used as a recombinant protein (61, 62), or specific peptides prepared from rFgCatL1 (63, 64). Using another test different than ELISA, the lateral flow test based on rFhCatL1showed maximal specificity and sensitivity compared to ELISA either against human serum or whole blood samples (65). Recently, the recombinant antigen of cathepsin L1 prepared from Fasciola strain wuh2-1 (F. hepatica/gigantica hybrid type) showed a high efficacy in diagnosis of fascioliasis using indirect ELISAs against sera obtained from Sika deer (66), and human sera (67) from Japan.
Thus, cathepsin L1 demonstrated its potential ability as a global diagnostic antigen for the detection of fascioliasis of various Fasciola species, in different diagnostic tests and for multiple susceptible species. The developed tests demonstrated sensitivity from 88% to a maximum of 100% and specificity from 94.6 to 100% in highly susceptible animals like sheep, goat, and cattle. However, these values have been slightly reduced in other hosts as horse and human. The results obtained are satisfactory, and therefore, some of the tests may overcome the disadvantages of the reference technique or become an efficient screening method. However, although indirect ELISAs demonstrate efficiency in the diagnosis of fascioliasis, other tests are used less frequently. Among these, western blotting and MFI are reliable methods with a high sensitivity and specificity for antibody detection and are widely used to evaluate serological assays performances. Even for ELISA, indirect ELISA format tests are mostly used, which are both sensitive and specific. Other ELISA formats should be also developed to increase the accuracy of this technique, for example, competitive ELISA, blocking ELISA, and capture ELISA, which use mAbs or pAbs that bind to the same epitope. Moreover, these tests do not require the use of secondary antibodies, which are specific for different species, and IgM and IgG antibodies can be tested in the same assay.
Most of the tests developed in the last decade have focused mainly in detection of antibodies against F. hepatica rather than F. gigantica and in human instead of highly susceptible animals as sheep and cattle (Table 2), and only very few studies focused on the serodiagnosis of fascioliasis in other species. In order to minimize false positives, most of the work done in the last few years has used specific Fasciola recombinant proteins (Table 2) using different expression systems. However, the use of native ES proteins in their original conformation avoids undesirable changes that can occur in heterologous expression systems (11). However, recombinant antigens have been used to attempt to overcome the disadvantage of variation in quality and quantity of native antigens derived directly from Fasciola. Also, the use of purified native antigens has a high probability of cross reactivity (47). Purification of proteins may also influence the performance of the developed tests. Thus, preparation of specific peptides based on epitopes mapping and selection of unique sequences and using as single peptide or as chimeric protein using available the free or the commercial online websites or programs will improve the diagnostic procedures against fascioliasis in animals and humans.
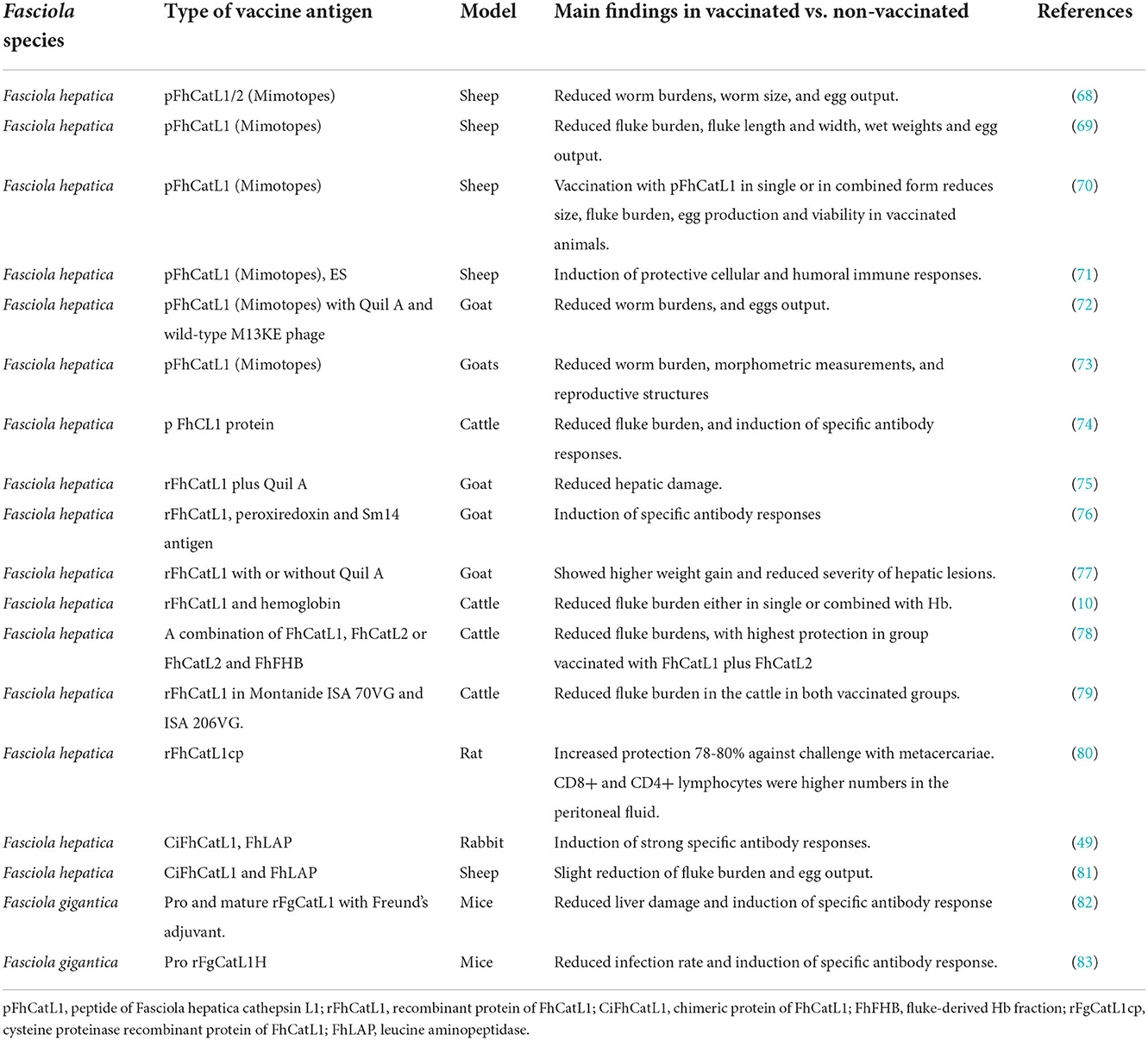
Similarly, numerous studies have assessed FhCatL1 and FgCatL1 as potential vaccine candidates using different animal models (Table 3). In this regard, several studies have demonstrated the immunoprotective properties of mimotopes derived from FhCatL1: Mimotopes are peptides mimicking protein that can be generated by phage display technology. This strategy showed promising results in vaccinated sheep as evidenced in reduced fluke burden and egg output combined with elicited protective immune response (68–71). Similar results were obtained when goats (72, 73), and cattle (74) were used as a model for vaccination using FhCatL1 mimotopes.
Furthermore, rFhCatL1 alone or formulated with Quil A adjuvant as a different approach of vaccination showed partial protection manifested in reduced liver changes and increased body gains in immunized goats (75–77) compared to the control non-immunized groups. Similar tendency was also reported when cattle was immunized with rFhCatL1 (10, 78, 79). Even in rat, enteral vaccination with rFhCatL1 induced high protection against challenge with fluke metacercariae (80).
Other studies have reported the usefulness of chimeric vaccine antigen prepared from FhCatL1 and leucine aminopeptidase (FhLAP) in eliciting an immune response and conferring protection against challenge infection in both rabbit (49), and sheep (81). Concerning vaccine researches applied on F. gigantica, immunization of mice with recombinant pro and mature FgCatL1 showed a protective potential against challenge infection when compared with non-immunized control group (82, 83).
These results revealed the usefulness of cathepsin L1 as a vaccine antigen against ovine, caprine, bovine and murine fascioliasis. Vaccines can be used to prevent, manage, or eradicate disease and are set to become increasingly important as front-line control tools, especially as many parasites including Fasciola progressively emerge with wide resistance to available anti-parasitic drugs and the burden of parasites. Different preparations of FhCatL1 including recombinant protein or mimotope and either used solitary or in combination with other antigens succeeded to confer efficient protective potentials in different animal models. However, further studies are required to improve the protective potentials, eg. by mixing with potent adjuvants, testing the underestimated F. gigantica, and finding out novel protective indices. As the vaccine preparation and testing are costly procedures, initial assessment of the efficacy will be highly advantageous in many aspects particularly from economic viewpoint. Bioinformatic analysis has been already providing this benefit for vaccine researches including those conducted for Fasciola assessments (20). The identification of potent immuno-epitopes from cathepsin L1 from F. hepatica or F. gigantica using our presented analyses software might contribute in improvement of vaccine efficacy via wet-lab approach not only the theoretical aspect. This could be proven via conducting certain preliminary experiments based on the interaction of a prepared antigen with immune-effectors cells (macrophages, dendritic cells, Treg cells) or molecules (Interleukin β1, IL-10, MHC classes I and II proteins) (84, 85). Currently, the selection of the targeted cell or molecule can be determined using a protein or peptide of known sequence via the available online websites and programs software.
Conclusion
We revealed the first description of multiple sequence alignment, isoelectric point, phosphorylation sites, and morpho-physical properties from protein database of FhCatL1. Moreover, first assessment of FhCatL1 was applied for secondary structure using PSIPRED and GOR IV servers, tertiary, and quaternary structure using SWISS-MODEL, and linear B- cell epitopes using ProtScale online available tool. The bioinformatic analyses in the present study revealed the high antigenicity, immunogenicity, hydrophilicity, surface accessibility, and flexibility indexes for FhCatL1 protein. Thus, application of on-line available bioinformatic analysis tools can be used for evaluation and validation of basic structures and properties and to predict antigenicity and immunogenicity of protein based on amino acid sequence. Such data are additionally supported with numerous previous literatures that corroborating the high efficiency of FhCatL1 as diagnostic and vaccine antigen. Hence, we recommend FhCatL1 as a suitable antigen for developing diagnostic tests and vaccines against fascioliasis in different animal models. Prediction of the antigenic or immunogenic properties of Fasciola- or other parasites-derived proteins via computational and online software programs will reduce the time, effort and cost before application of practical assays and experiments. Useful relevant information was also presented for FgCatL1 as a diagnostic and vaccine candidates, although it was much less than those of FhCatL1.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Author contributions
Conceptualization and investigation: RF and AA. Formal analysis, project administration, and writing—original draft: RF. Methodology: RF, SM, E-SE-A, HA, and MO. Resources and writing—review and editing: RF, SM, E-SE-A, HA, OS, MO, and AA. All authors contributed to the article and approved the submitted version.
Acknowledgments
The researchers would like to thank the Deanship of Scientific Research, Qassim University for funding the publication of this project.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpubh.2022.1054502/full#supplementary-material
References
1. Köstenberger K, Tichy A, Bauer K, Pless P, Wittek T. Associations between fasciolosis and milk production, and the impact of anthelmintic treatment in dairy herds. Parasitol Res. (2017) 116:1981–7. doi: 10.1007/s00436-017-5481-3
2. Charlier J, Hostens M, Jacobs J, Van Ranst B, Duchateau L, Vercruysse J, et al. Integrating fasciolosis control in the dry cow management: the effect of closantel treatment on milk production. PLoS ONE. (2021) 7:e43216. doi: 10.1371/journal.pone.0043216
3. Amer S, ElKhatam A, Zidan S, Feng Y, Xiao L. Identity of Fasciola spp. in sheep in Egypt. Parasit Vectors. (2016) 9:623. doi: 10.1186/s13071-016-1898-2
4. Fox NJ, White PC, McClean CJ, Marion G, Evans A, Hutchings MR, et al. Predicting impacts of climate change on Fasciola hepatica risk. PLoS ONE. (2011) 6:e16126. doi: 10.1371/journal.pone.0016126
5. Kamaludeen J, Graham-Brown J, Stephens N, Miller J, Howell A, Beesley NJ, et al. Lack of efficacy of triclabendazole against Fasciola hepatica is present on sheep farms in three regions of England, and Wales. Vet Rec. (2019) 184:502. doi: 10.1136/vr.105209
6. Charlier J, Vercruysse J, Morgan E, van Dijk J, Williams DJ. Recent advances in the diagnosis, impact on production and prediction of Fasciola hepatica in cattle. Parasitology. (2014) 141:326–35. doi: 10.1017/S0031182013001662
7. Salimi-Bejestani MR, McGarry JW, Felstead S, Ortiz P, Akca A, Williams DJ, et al. Development of an antibody-detection ELISA for Fasciola hepatica and its evaluation against a commercially available test. Res Vet Sci. (2005) 78:177–81. doi: 10.1016/j.rvsc.2004.08.005
8. Maggio D, Tirloni LS, Pinto L, Diedrich AF, Yates Iii JK, Benavides U, et al. IJr; Berasain, P Across intra-mammalian stages of the liver f luke Fasciola hepatica: a proteomic study Sci Rep. (2016) 6:32796. doi: 10.1038/srep32796
9. Robinson MW, Tort JF, Lowther J, Donnelly SM, Wong E, Xu W, et al. Proteomics and phylogenetic analysis of the cathepsin L protease family of the helminth pathogen Fasciola hepatica: expansion of a repertoire of virulence-associated factors. Mol Cell Proteomics: MCP. (2008) 7:1111–23. doi: 10.1074/mcp.M700560-MCP200
10. Dalton JP, McGonigle S, Rolph TP, Andrews SJ. Induction of protective immunity in cattle against infection with Fasciola hepatica by vaccination with cathepsin L proteinases and with hemoglobin. Infect Immun. (1996) 64:5066–74. doi: 10.1128/iai.64.12.5066-5074.1996
11. Alvarez Rojas CA, Jex AR, Gasser RB, Scheerlinck JP. Techniques for the diagnosis of Fasciola infections in animals: room for improvement. Adv Parasitol. (2014) 85:65–107. doi: 10.1016/B978-0-12-800182-0.00002-7
12. Collett CF, Phillips HC, Fisher M, Smith S, Fenn C, Goodwin P, et al. Fasciola hepatica cathepsin L zymogens: immuno-proteomic evidence for highly immunogenic zymogen-specific conformational epitopes to support diagnostics development. J Proteome Res. (2022) 21:1997–2010. doi: 10.1021/acs.jproteome.2c00299
13. Romano P, Giugno R, Pulvirenti A. Tools collaborative environments for bioinformatics research. Brief Bioinformatics. (2011) 12:549–61. doi: 10.1093/bib/bbr055
14. Van Regenmortel MH. Protein structure and antigenicity. Int J Rad Appl Instrum B. (1987) 14:277–80. doi: 10.1016/0883-2897(87)90003-1
15. Debelle L, Wei S, Jacob M, Hornebeck W, Alix A. Predictions of the secondary structure and antigenicity of human and bovine tropoelastins. Eur Biophys J. (1992) 21:321–9. doi: 10.1007/BF00188344
16. Van Regenmortel MH. Which structural features determine protein antigenicity? Trends Biochem Sci. (1986) 11:36–9. doi: 10.1016/0968-0004(86)90230-6
17. Wang Y, Wu W, Negre NN, White KP, Li C, Shah PK, et al. Determinants of antigenicity and specificity in immune response for protein sequences. BMC Bioinformatics. (2011) 12:1–13. doi: 10.1186/1471-2105-12-251
19. Aghamolaei S, Kazemi B, Bandehpour M, Ranjbar MM, Rouhani S, Javadi Mamaghani A, et al. Design and expression of polytopic construct of cathepsin-L1, SAP-2 and FhTP16.5 proteins of Fasciola hepatica. J. Helminthol. (2020) 94:e134. doi: 10.1017/S0022149X20000140
20. Akil M, Aykur M, Karakavuk M, Can H, Döşkaya M. Construction of a multiepitope vaccine candidate against Fasciola hepatica: an in silico design using various immunogenic excretory/secretory antigens. Expert Rev Vaccines. (2021) 1–14. doi: 10.1080/14760584.2022.1996233
21. Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods. (2011) 8:785–786. doi: 10.1038/nmeth.1701
22. Garnier J, Gibrat JF, Robson BGOR. method for predicting protein secondary structure from amino acid sequence. Meth Enzymol. (1996) 266:540–53. doi: 10.1016/S0076-6879(96)66034-0
23. Buchan D, Jones DT. The PSIPRED protein analysis workbench: 20 years on. Nucleic Acids Res. (2019) 47:W402–7. doi: 10.1093/nar/gkz297
24. Guex N, Peitsch MC, Schwede T. Automated comparative protein structure modeling with SWISS-MODEL and Swiss-PdbViewer: a historical perspective. Electrophoresis. (2009) 30:S162–73. doi: 10.1002/elps.200900140
25. Gasteiger E, Hoogland C, Gattiker A, Wilkins MR, Appel RD, Bairoch A. Protein identification and analysis tools on the ExPASy server. In: The Proteomics Protocols Handbook, Springer. (2005) p. 571–607. doi: 10.1385/1-59259-890-0:571
26. Parker J, Guo D, Hodges R. New hydrophilicity scale derived from high-performance liquid chromatography peptide retention data: correlation of predicted surface residues with antigenicity and X-ray-derived accessible sites. Biochemistry. (1986) 25:5425–32. doi: 10.1021/bi00367a013
27. Kolaskar A, Tongaonkar PCA. semi-empirical method for prediction of antigenic determinants on protein antigens. FEBS Lett. (1990) 276:172–4. doi: 10.1016/0014-5793(90)80535-Q
28. Cornelissen JB, Gaasenbeek CP, Borgsteede FH, Holland WG, Harmsen MM, Boersma WJ, et al. Early immunodiagnosis of fasciolosis in ruminants using recombinant Fasciola hepatica cathepsin L-like protease. Int J Parasitol. (2001) 31:728–37. doi: 10.1016/S0020-7519(01)00175-8
29. Shapovalov G, Ritaine A, Bidaux G, Slomianny C, Borowiec AS, Gordienko D, et al. Organelle membrane derived patches: reshaping classical methods for new targets. Sci Rep. (2017) 7:14082. doi: 10.1038/s41598-017-13968-y
31. Reuven EM, Fink A, Shai Y. Regulation of innate immune responses by transmembrane interactions: lessons from the TLR family. Biochim Biophys Acta. (2014) 1838:1586–93. doi: 10.1016/j.bbamem.2014.01.020
32. Kozlowski LPIPC. Isoelectric point calculator. Biol Direct. (2011) 11:55. doi: 10.1186/s13062-016-0159-9
33. Hansson M, Nygren PA, Ståhl S. Design production of recombinant subunit vaccines. Biotechnol Appl Biochem. (2000) 32:95–107. doi: 10.1042/BA20000034
34. Walsh CP. Osttranslational Modification of Proteins: Expanding Nature's Inventory. Greenwood, CO: Roberts and Company Publishers (2006).
35. Yada RY, Jackman RL, Nakai S. Secondary structure prediction and determination of proteins: a review. Int J Pept Protein Res. (1988) 31:98–108. doi: 10.1111/j.1399-3011.1988.tb00011.x
36. Chen VB, Arendall WB, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D. (2010) 66:12–21. doi: 10.1107/S0907444909042073
37. Benkert P, Biasini M, Schwede T. Toward the estimation of the absolute quality of individual protein structure models. Bioinformatics. (2011) 27:343–50. doi: 10.1093/bioinformatics/btq662
38. Hochleitner EO, Sondermann P, Lottspeich F. Determination of the stoichiometry of protein complexes using liquid chromatography with fluorescence and mass spectrometric detection of fluorescently labeled proteolytic peptides. Proteomics. (2004) 4:669–76. doi: 10.1002/pmic.200300668
39. Scheiblhofer S, Laimer J, Machado Y, Weiss R, Thalhamer J. Influence of protein fold stability on immunogenicity and its implications for vaccine design. Expert Rev Vaccines. (2017) 16:479–89. doi: 10.1080/14760584.2017.1306441
40. Kringelum JV, Lundegaard C, Lund O, Nielsen M. Reliable B cell epitope predictions: impacts of method development and improved benchmarking. PLoS Comput Biol. (2012) 8:e1002829. doi: 10.1371/journal.pcbi.1002829
41. Bijker MS, Melief CJ, Offringa R, Burg SH. Design and development of synthetic peptide vaccines: past, present and future. Expert Rev Vaccines. (2007) 6:591–603. 10.1586/14760584.6.4.591 doi: 10.1586/14760584.6.4.591
42. Gomara MJ, Haro I. Synthetic peptides for the immunodiagnosis of human diseases. Curr Med Chem. (2007) 14:531–546. doi: 10.2174/092986707780059698
43. Martínez-Sernández V, Perteguer MJ, Hernández-González A, Mezo M, González-Warleta M, Orbegozo-Medina RA, et al. Comparison of recombinant cathepsins L1, L2, and L5 as ELISA targets for serodiagnosis of bovine and ovine fascioliasis. Parasitol Res. (2018) 117:1521–34. doi: 10.1007/s00436-018-5809-7
44. Mokhtarian K, Meamar AR, Khoshmirsafa M, Razmjou E, Masoori L, Khanmohammadi M, et al. Comparative assessment of recombinant and native immunogenic forms of Fasciola hepatica proteins for serodiagnosis of sheep fasciolosis. Parasitol Res. (2018) 117:225–32. doi: 10.1007/s00436-017-5696-3
45. Walsh TR, Ainsworth S, Armstrong S, Hodgkinson J, Williams D. Differences in the antibody response to adult Fasciola hepatica excretory/secretory products in experimentally and naturally infected cattle and sheep. Vet Parasitol. (2021) 289:109321. doi: 10.1016/j.vetpar.2020.109321
46. Kuerpick B, Schnieder T, Strube C. Evaluation of a recombinant cathepsin L1 ELISA and comparison with the Pourquier and ES ELISA for the detection of antibodies against Fasciola hepatica. Vet Parasitol. (2013) 193:206–13. doi: 10.1016/j.vetpar.2012.11.021
47. Cornelissen JB, Gaasenbeek CP, Boersma W, Borgsteede FH, van Milligen FJ. Use of a pre-selected epitope of cathepsin-L1 in a highly specific peptide-based immunoassay for the diagnosis of Fasciola hepatica infections in cattle. Int J Parasitol. (1999) 29:685–96. doi: 10.1016/S0020-7519(99)00017-X
48. Karanikola SN, Krücken J, Ramünke S, Waal d. e., Höglund T, Charlier J, et al. of a multiplex fluorescence immunological assay for the simultaneous detection of antibodies against Cooperia oncophora, Dictyocaulus viviparus and Fasciola hepatica in cattle. Parasit Vectors. (2015) 8:335. doi: 10.1186/s13071-015-0924-0
49. Hernández-Guzmán K, Sahagún-Ruiz A, Vallecillo AJ, Cruz-Mendoza I, Quiroz-Romero H. Construction evaluation of a chimeric protein made from Fasciola hepatica leucine aminopeptidase and cathepsin L1. J Helminthol. (2016) 90:7–13. doi: 10.1017/S0022149X14000686
50. Quigley A, Sekiya M, Egan S, Wolfe A, Negredo C, Mulcahy G, et al. (2017). Prevalence of liver fluke infection in Irish horses and assessment of a serological test for diagnosis of equine fasciolosis. Equine Vet. 49:183–188. doi: 10.1111/evj.12577
51. Raina OK, Yadav SC, Sriveny D, Gupta SC. Immuno-diagnosis of bubaline fasciolosis with Fasciola gigantica cathepsin-L and recombinant cathepsin L 1-D proteases. Acta Trop. (2006) 98:145–51. doi: 10.1016/j.actatropica.2006.03.004
52. Meshgi B, Jalousian F, Fathi S, Jahani Z. Design and synthesis of a new peptide derived from Fasciola gigantica cathepsin L1 with potential application in serodiagnosis of fascioliasis. Exp Parasitol. (2018) 189:76–86. doi: 10.1016/j.exppara.2018.04.013
53. Anuracpreeda P, Chawengkirttikul R, Sobhon P. Immunodiagnosis of Fasciola gigantica infection using monoclonal antibody-based sandwich ELISA and immunochromatographic assay for detection of circulating cathepsin L1 protease. PLoS ONE. (2016) 11:e0145650. doi: 10.1371/journal.pone.0145650
54. O'Neill SM, Parkinson M, Strauss W, Angles R, Dalton JP. Immunodiagnosis of Fasciola hepatica infection (fascioliasis) in a human population in the Bolivian Altiplano using purified cathepsin L cysteine proteinase. Am J Trop Med Hyg. (1998) 58:417–23. doi: 10.4269/ajtmh.1998.58.417
55. O'Neill SM, Parkinson M, Dowd AJ, Strauss W, Angles R, Dalton JP, et al. Short report: Immunodiagnosis of human fascioliasis using recombinant Fasciola hepatica cathepsin L1 cysteine proteinase. Am J Trop Med Hyg. (1999) 60:749–51. doi: 10.4269/ajtmh.1999.60.749
56. Carnevale S, Rodríguez MI, Guarnera EA, Carmona C, Tanos T, Angel SO, et al. Immunodiagnosis of fasciolosis using recombinant procathepsin L cystein proteinase. Diagn Microbiol Infect Dis. (2001) 41:43–9. doi: 10.1016/S0732-8893(01)00288-7
57. Rokni MB, Massoud J, O'Neill SM, Parkinson M, Dalton JP. Diagnosis of human fasciolosis in the Gilan province of Northern Iran: application of cathepsin L-ELISA. Diagn Microbiol Infect Dis. (2002) 44:175–9. doi: 10.1016/S0732-8893(02)00431-5
58. Gonzales Santana B, Dalton JP, Vasquez Camargo F, Parkinson M, Ndao M. The diagnosis of human fascioliasis by enzyme-linked immunosorbent assay (ELISA) using recombinant cathepsin L protease. PLoS Negl Trop Dis. (2013) 7:e2414. doi: 10.1371/journal.pntd.0002414
59. Gottstein B, Schneeberger M, Boubaker G, Merkle B, Huber C, Spiliotis M, et al. Comparative assessment of ELISAs using recombinant saposin-like protein 2 and recombinant cathepsin L-1 from Fasciola hepatica for the serodiagnosis of human Fasciolosis. PLoS Negl Trop Dis. (2014) 8:e2860. doi: 10.1371/journal.pntd.0002860
60. Mokhtarian K, Akhlaghi L, Mohammadi M, Meamar AR, Razmjou E, Khoshmirsafa M, et al. Evaluation of anti-Cathepsin L1: a more reliable method for serodiagnosis of human fasciolosis. Trans R Soc Trop Med Hyg. (2016) 110:542–50. doi: 10.1093/trstmh/trw063
61. Tantrawatpan C, Maleewong W, Wongkham C, Wongkham S, Intapan PM, Nakashima K, et al. Serodiagnosis of human fascioliasis by a cystatin capture enzyme-linked immunosorbent assay with recombinant Fasciola gigantica cathepsin L antigen. Am J Trop Med Hyg. (2005) 72:82–6. doi: 10.4269/ajtmh.2005.72.82
62. Wongkham C, Tantrawatpan C, Intapan PM, Maleewong W, Wongkham S, Nakashima K, et al. Evaluation of immunoglobulin G subclass antibodies against recombinant Fasciola gigantica cathepsin L1 in an enzyme-linked immunosorbent assay for serodiagnosis of human fasciolosis. Clin Diagn Lab Immunol. (2005) 12:1152–6. doi: 10.1128/CDLI.12.10.1152-1156.2005
63. Intapan PM, Tantrawatpan C, Maleewong W, Wongkham S, Wongkham C, Nakashima K, et al. Potent epitopes derived from Fasciola gigantica cathepsin L1 in peptide-based immunoassay for the serodiagnosis of human fascioliasis. Diagn Microbiol Infect Dis. (2005) 53:125–9. doi: 10.1016/j.diagmicrobio.2005.05.010
64. Tantrawatpan C, Maleewong W, Wongkham C, Wongkham S, Intapan PM, Nakashima K, et al. Evaluation of immunoglobulin G4 subclass antibody in a peptide-based enzyme-linked immunosorbent assay for the serodiagnosis of human fascioliasis. Parasitology. (2007) 134:2021–6. doi: 10.1017/S0031182007003435
65. Martínez-Sernández V, Muiño L, Perteguer MJ, Gárate T, Mezo M, González-Warleta M, et al. Development and evaluation of a new lateral flow immunoassay for serodiagnosis of human fasciolosis. PLoS Neg Trop Dis. (2011) 5:e1376. doi: 10.1371/journal.pntd.0001376
66. Sato H, Hiraya H, Sugiyama T, Fukumoto S, Matsuyama R, Yanagawa Y, et al. Seroprevalence of fasciolosis in Hokkaido sika deer (Cervus nippon yesoensis) from Hokkaido Prefecture, Japan revealed by ELISA using recombinant cathepsin L1. Parasitol Int. (2021) 80:102222. doi: 10.1016/j.parint.2020.102222
67. Sugiyama T, Ichikawa-Seki M, Sato H, Kounosu A, Tanaka M, Maruyama H, et al. Enzyme-linked immunosorbent assay (ELISA) using recombinant Fasciola cathepsin L1 for the diagnosis of human fasciolosis caused by Fasciola hepatica/gigantica hybrid type. Parasitol Int. (2021) 82:102311. doi: 10.1016/j.parint.2021.102311
68. Villa-Mancera A, Quiroz-Romero H, Correa D, Ibarra F, Reyes-Pérez M, Reyes-Vivas H, et al. Induction of immunity in sheep to Fasciola hepatica with mimotopes of cathepsin L selected from a phage display library. Parasitology. (2008) 135:1437–45. doi: 10.1017/S003118200800471X
69. Villa-Mancera A, Méndez-Mendoza M. Protection antibody isotype responses against Fasciola hepatica with specific antibody to pIII-displayed peptide mimotopes of cathepsin L1 in sheep. Vet J. (2012) 194:108–12. doi: 10.1016/j.tvjl.2012.03.003
70. Villa-Mancera A, Alcalá-Canto Y, Reynoso-Palomar A, Olmedo-Juárez A, Olivares-Pérez J. Vaccination with cathepsin L phage-exposed mimotopes, single or in combination, reduce size, fluke burden, egg production and viability in sheep experimentally infected with Fasciola hepatica. Parasitol. Int. (2021) 83:102355. doi: 10.1016/j.parint.2021.102355
71. Villa-Mancera A, Olivares-Pérez J, Olmedo-Juárez A, Reynoso-Palomar A. Phage display-based vaccine with cathepsin L and excretory-secretory products mimotopes of Fasciola hepatica induces protective cellular and humoral immune responses in sheep. Vet Parasitol. (2021) 289:109340. doi: 10.1016/j.vetpar.2020.109340
72. Villa-Mancera A, Reynoso-Palomar A, Utrera-Quintana F, Carreón-Luna L. Cathepsin L1 mimotopes with adjuvant Quil A induces a Th1/Th2 immune response and confers significant protection against Fasciola hepatica infection in goats. Parasitol Res. (2014) 113:243–50. doi: 10.1007/s00436-013-3650-6
73. Villa-Mancera A, Alcalá-Canto Y, Olivares-Pérez J, Molina-Mendoza P, Hernández-Guzmán K, Utrera-Quintana F, et al. Vaccination with cathepsin L mimotopes of Fasciola hepatica in goats reduces worm burden, morphometric measurements, reproductive structures. Microb Pathog. (2021) 155:104859. doi: 10.1016/j.micpath.2021.104859
74. Garza-Cuartero L, Geurden T, Mahan SM, Hardham JM, Dalton JP, Mulcahy G, et al. Antibody recognition of cathepsin L1-derived peptides in Fasciola hepatica-infected and/or vaccinated cattle and identification of protective linear B-cell epitopes. Vaccine. (2018) 36:958–968. doi: 10.1016/j.vaccine.2018.01.020
75. Pérez-Ecija RA, Mendes RE, Zafra R, Buffonni L, Martínez-Moreno A, Pérez J, et al. Pathological and parasitological protection in goats immunised with recombinant cathepsin L1 and challenged with Fasciola hepatica. Vet J. (2010) 185:351–3. doi: 10.1016/j.tvjl.2009.07.004
76. Buffoni L, Martínez-Moreno FJ, Zafra R, Mendes RE, Pérez-Écija A, Sekiya M, et al. Humoral immune response in goats immunised with cathepsin L1, peroxiredoxin and Sm14 antigen and experimentally challenged with Fasciola hepatica. Vet Parasitol. (2012) 185:315–21. doi: 10.1016/j.vetpar.2011.09.027
77. Zafra R, Pérez-Écija RA, Buffoni L, Moreno P, Bautista MJ, Martínez-Moreno A, et al. Early and late peritoneal and hepatic changes in goats immunized with recombinant cathepsin L1 and infected with Fasciola hepatica. J Comp Pathol. (2013) 148:373–84. doi: 10.1016/j.jcpa.2012.08.007
78. Mulcahy G, O'Connor F, Clery D, Hogan SF, Dowd AJ, Andrews SJ, et al. Immune responses of cattle to experimental anti-Fasciola hepatica vaccines. Res Vet Sci. (1999) 67:27–33. doi: 10.1053/rvsc.1998.0270
79. Golden O, Flynn RJ, Read C, Sekiya M, Donnelly SM, Stack C, et al. Protection of cattle against a natural infection of Fasciola hepatica by vaccination with recombinant cathepsin L1 (rFhCL1). Vaccine. (2010) 28:5551–7. doi: 10.1016/j.vaccine.2010.06.039
80. Kesik M, Jedlina-Panasiuk L, Kozak-Cieszczyk M, Płucienniczak A, Wedrychowicz H. Enteral vaccination of rats against Fasciola hepatica using recombinant cysteine proteinase (cathepsin L1). Vaccine. (2007) 25:3619–28. doi: 10.1016/j.vaccine.2007.01.057
81. Ortega-Vargas S, Espitia C, Sahagún-Ruiz A, Parada C, Balderas-Loaeza A, Villa-Mancera A, et al. Moderate protection is induced by a chimeric protein composed of leucine aminopeptidase and cathepsin L1 against Fasciola hepatica challenge in sheep. Vaccine. (2019) 37:3234–40. doi: 10.1016/j.vaccine.2019.04.067
82. Kueakhai P, Changklungmoa N, Chaichanasak P, Jaikua W, Itagaki T, Sobhon P, et al. Vaccine potential of recombinant pro- and mature cathepsinL1 against fasciolosis gigantica in mice. Acta Trop. (2015) 150:71–78. doi: 10.1016/j.actatropica.2015.06.020
83. Sansri V, Meemon K, Changklungmoa N, Kueakhai P, Chantree P, Chaichanasak P, et al. Protection against Fasciola gigantica infection in mice by vaccination with recombinant juvenile-specific cathepsin L. Vaccine. (2015) 33:1596–601. doi: 10.1016/j.vaccine.2015.02.010
84. Moreau E, Chauvin A. Immunity against helminths: interactions with the host and the intercurrent infections. J Biomed Biotechnol. (2010) 2010:428593. doi: 10.1155/2010/428593
Keywords: Fasciola hepatica, antigen, diagnosis, vaccine, cathepsin L1
Citation: Fereig RM, Metwally S, El-Alfy E-S, Abdelbaky HH, Shanab O, Omar MA and Alsayeqh AF (2022) High relatedness of bioinformatic data and realistic experimental works on the potentials of Fasciola hepatica and F. gigantica cathepsin L1 as a diagnostic and vaccine antigen. Front. Public Health 10:1054502. doi: 10.3389/fpubh.2022.1054502
Received: 26 September 2022; Accepted: 17 November 2022;
Published: 07 December 2022.
Edited by:
Ana Afonso, Universidade NOVA de Lisboa, PortugalCopyright © 2022 Fereig, Metwally, El-Alfy, Abdelbaky, Shanab, Omar and Alsayeqh. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Abdullah F. Alsayeqh, a.alsayeqh@qu.edu.sa
 Ragab M. Fereig
Ragab M. Fereig Samy Metwally2
Samy Metwally2 El-Sayed El-Alfy
El-Sayed El-Alfy Hanan H. Abdelbaky
Hanan H. Abdelbaky Mosaab A. Omar
Mosaab A. Omar Abdullah F. Alsayeqh
Abdullah F. Alsayeqh