- 1Department of Genomics and Biology of Fruit Crops, Research and Innovation Centre, Fondazione Edmund Mach (FEM), San Michele all'Adige, Italy
- 2Laboratory of Biotechnology and Plant Pathology, DISTAL, University of Bologna, Bologna, Italy
- 3Plant Protection Research Division of Melkassa Agricultural Research Center, Ethiopian Institute of Agricultural Research (EIAR), Addis Ababa, Ethiopia
- 4Unit of Computational Biology, Research and Innovation Centre, Fondazione Edmund Mach (FEM), San Michele all'Adige, Italy
- 5Department of Food Quality and Nutrition, Research and Innovation Centre, Fondazione Edmund Mach, San Michele all'Adige, Italy
- 6ESAT-ELECTA, Electrical Energy and Computer Architectures, Leuven, Belgium
Botrytis cinerea is an important necrotroph in vineyards. Primary infections are mostly initiated by airborne conidia from overwintered sources around bloom, then the fungus remains quiescent from bloom till maturity and egresses at ripeness. We previously described in detail the process of flower infection and quiescence initiation. Here, we complete the characterization studying the cross-talk between the plant and the fungus during pathogen quiescence and egression by an integrated transcriptomic and metabolic analysis of the host and the pathogen. Flowers from fruiting cuttings of the cv. Pinot Noir were inoculated with a GFP-labeled strain of B. cinerea at full cap-off stage, and molecular analyses were carried out at 4 weeks post inoculation (wpi, fungal quiescent state) and at 12 wpi (fungal pre-egression and egression states). The expressed fungal transcriptome highlighted that the fungus remodels its cell wall to evade plant chitinases besides undergoing basal metabolic activities. Berries responded by differentially regulating genes encoding for different PR proteins and genes involved in monolignol, flavonoid, and stilbenoid biosynthesis pathways. At 12 wpi, the transcriptome of B. cinerea in the pre-egressed samples showed that virulence-related genes were expressed, suggesting infection process was initiated. The egressed B. cinerea expressed almost all virulence and growth related genes that enabled the pathogen to colonize the berries. In response to egression, ripe berries reprogrammed different defense responses, though futile. Examples are activation of membrane localized kinases, stilbene synthases, and other PR proteins related to SA and JA-mediated responses. Our results indicated that hard-green berries defense program was capable to hamper B. cinerea growth. However, ripening associated fruit cell wall self-disassembly together with high humidity created the opportunity for the fungus to egress and cause bunch rot.
Introduction
Botrytis cinerea is a necrotrophic fungus responsible for significant economic losses in vineyards by causing bunch rot. The disease is mostly observed on ripe berries, following rainfalls or a long period of high humidity close to harvest, and develops into gray mold. Primary infections are usually initiated by airborne conidia from overwintered sources (Nair et al., 1995; Elmer and Michailides, 2004) and mostly occur at bloom leading to quiescent infection (McClellan and Hewitt, 1973; Keller et al., 2003; Pezet et al., 2003b). Quiescent infection is an interesting phenomenon in B. cinerea–plant interaction where the pathogen spends prolonged time in the host tissue asymptomatically, without being aggressive (Williamson et al., 1987; McNicol and Williamson, 1989; Coertze and Holz, 2002; Shaw et al., 2016). Recently, it has been observed in grapevine that B. cinerea inoculated at full bloom stays quiescent until full maturity and egresses at ripening causing bunch rot (Haile et al., 2017; Haile et al., 2019).
What drives and keeps B. cinerea into quiescence until berry ripening is not fully known, but preformed and induced defense mechanisms, including immature berries skin features such as polyphenols in the berry skin cell wall and the thickness of the epidermal cell layer complex, have been proposed as part of the ontogenic resistance to B. cinerea (Goetz et al., 1999; Keller et al., 2003; Deytieux-Belleau et al., 2009). Molecular analysis has shown that upon contact with the grapevine flower, B. cinerea induces genes, both encoding known virulence factors, such as boctinic acid (BcBOA6), botrydial phytotoxin (BcBOT1 and BcBOT2), polygalacturonase 2 (BcPG2), and superoxide dismutase 1 (BcSOD1) encoding genes, and contributing to the infection program, such as oxaloacetate acetyl hydrolase (BcOAH), endo-β-1,4-xylanase (BcXYN11A), and glutathione S-transferase (BcGST1) encoding genes, to cause disease (Haile et al., 2017). However, no visible disease progress was observed despite the confirmed presence of the pathogen on the immature berries (Haile et al., 2017). As a response to the infection attempt, grapevine flowers react by reprogramming the expression of genes encoding antimicrobial proteins, monolignol biosynthesis (VvPAL, VvCOMT, VvCCoAMT, and VvCAD), stilbenoids (VvSTSs), and prompting oxidative burst (VvGLP3). These induced defense responses of the grapevine flowers are presumably involved in B. cinerea quiescence (Haile et al., 2017). Another study highlighted the involvement of the salicylic acid (SA) dependent defense pathway together with the accumulation of ROS and the activation of stilbene and lignin biosynthesis as main factors arresting B. cinerea progress on véraison berries but not in the ripe ones, that were fully susceptible to the pathogen (Kelloniemi et al., 2015).
The transition from a quiescent to an active infection mostly occurs during fruit ripening. Physiological and biochemical changes that occur in the fruit during ripening, together with favorable climatic conditions during ripening, are suggested to trigger the transition (Prusky, 1996; Barnes and Shaw, 2002; Prusky et al., 2013). Cell wall loosening and appearance of disassembled cell wall substrates (Cantu et al., 2008), decrease in preformed and inducible host defense responses and change in hormonal balance and pH (Prusky, 1996; Prusky et al., 2013) are the major events during berry ripening that could enhance the egression and outgrowth of a quiescent necrotrophic pathogen. Egression impairs product quantity, quality, and appearance.
Global expression profiling of both pathogen and host at quiescent and egression stages of the infection enables to gain insights into signaling, metabolic pathways, transcriptional control, and defense responses involved in the cross-talk. Here we report the simultaneous transcriptome and secondary metabolite analyses of the B. cinerea–grapevine pathosystem, at hard-green and ripe stages, after host inoculation with B. cinerea conidia at full cap-off stage. Our results revealed that grapevine berries were able to keep the fungus quiescent for 12 weeks upon flower inoculation activating defense responses similar to the ones activated at bloom (Haile et al., 2017). On the other side, the pathogen was able to maintain its basal metabolic activities during quiescence and cause disease when the fruit activates some physiological responses which favor its egression, i.e. at ripening. These new molecular evidences represent a valuable resource in order to define the most appropriate infection stages for treatments against B. cinerea.
Materials and Methods
Fungal Isolate, Plant Material, and Inoculation
A genetically transformed B. cinerea strain, B05.10, expressing a green fluorescent protein (GFP) was used as in Haile et al., 2017. Grapevine fruiting cuttings obtained from Pinot Noir winter woody cuttings were grown and infected as described in Haile et al., 2017.
Flowers at full cap–fall stage [EL25/26, according to Eichorn and Lorenz (1977)] were inoculated by placing a 1.5 µl droplet of either conidia solution of GFP-labeled B05.10 (2*105 ml–1) or distilled water (mock inoculation) close to the receptacle area. Conidia were obtained from B. cinerea grown on PDA at 25°C for 10 days, and the concentration was determined using a hemacytometer under light microscope. Inoculation was made on three biological replicates, considering the inflorescence from a fruiting cutting as one biological replicate. After inoculation, the whole pot was immediately bagged in a clear plastic bag sprayed with water, for 24 h, in order to ensure high humidity around the inoculated inflorescence, which is an essential factor for conidial germination. Inoculated inflorescences were regularly inspected for gray mold growth until fruit ripening. At full coloring (approximately 10 weeks post inoculation, wpi), bunches were bagged for 2 weeks with plastic bags, to create favorable humidity for B. cinerea to egress.
Samples were collected at two time points, at 4 wpi (hard green berries), and at 12 wpi (ripe berries) when Botrytis egression was evident on a subset of berries. For the latter time point, two kinds of samples were collected: berries with visible egressed Botrytis and berries without visible Botrytis sign from a cluster. Samples without visible Botrytis signs are hereafter called berries with “pre-egressed” Botrytis, while the others are called berries with “egressed” Botrytis. Samples were snap frozen in liquid nitrogen and stored at -80 °C until use. For transcriptome and polyphenol analyses, fleshy exocarp was excised from individual fruits at the site of inoculation or where Botrytis symptoms were visible.
RNA Extraction, RNA-Seq, and qPCR Analyses
Extraction of RNA was performed as described in Haile et al., 2017. Single-end reads of 100 bp long sequences were obtained for each sample using a Next Generation Sequencing Platform HiSeq 1500 (Illumina, San Diego, CA). Approximately 20 million strand-specific sequences were obtained, except for pre-egressed samples (above 45 million), where the sequence depth was doubled in order to obtain more reads of Botrytis origin. The quality of the reads was checked using FastQC (version 0.11.2) software and pre-processed by cutadapt [version 1.8.1; Martin (2011)] for adapter. Genome assemblies of grapevine (12Xv1, http://genomes.cribi.unipd.it/) and B. cinerea (strain B05.10) (ASM83294v1, http://fungi.ensembl.org) were used as reference sequences. The alignment was made by Subread aligner (Liao et al., 2013) and raw read counts were extracted using the featureCount read summarization program (Liao et al., 2014). All raw RNA-Seq read data are deposited in the NCBI Short Read Archive (http://www.ncbi.nlm.nih.gov/sra/) under the BioProject accession code PRJNA414966.
The RNA sequences of B. cinerea (B05.10), from the PDB cultured conidia (used in Haile et al., 2017), were also used in this study as a control for determining the in planta Botrytis transcripts.
Finally, cDNA synthesis and quantitative polymerase chain reaction (qPCR) assay were carried out as described in Haile et al., 2017. For qPCR assay, each amplification reaction was run in triplicate, and VvACT and VvGAPDH, and BcRPL5 and BcTUBA genes were selected using GeNORM (Vandesompele et al., 2002) as reference genes for grapevine and B. cinerea gene expression normalization, respectively. Amplification efficiencies of each primer pair were calculated with LinReg software (Ruijter et al., 2009). The obtained amplification efficiency was used to calculate the relative quantity (RQ) and normalized relative quantity (NRQ) according to Hellemans et al. (2007). All primers and corresponding gene identifiers are listed in Supplemental Table S1.
Polyphenol Extraction and Analysis
Extraction of polyphenol and ultra high performance liquid chromatography–diode array detection–mass spectrometry (UHPLC–DAD–MSMS) analysis were carried out as described in Haile et al., 2017.
Statistical Analysis
qPCR Data
Statistical analyses of the qPCR results were made after log2(NRQ) transformation (Rieu and Powers, 2009). Statistical significance was calculated by Tukey’s honestly significant difference test or an unpaired heteroscedastic Student’s t test, considering each technical replicate as an individual sample.
RNA-Seq Data
For differential expression analysis, the voom method (Law et al., 2014) was applied to estimate the mean–variance relationship of the log-counts and produce a precision weight for each observation that was fed into the limma empirical Bayes analysis pipeline.(Smyth, 2004). Two-sample t-test was used for transcripts of grapevine at 4 wpi (mock inoculated vs Botrytis inoculated) and B. cinerea at egression (PDB-cultured Botrytis vs egressed Botrytis), whereas one-way ANOVA for grapevine transcripts at 12 wpi (mock inoculated vs pre-egressed Botrytis vs egressed Botrytis). Genes were considered differentially expressed (DE) if they fulfill a p-value of < 0.01 and an absolute fold change of ≥ 2.0.
Principal component analysis (PCA) was performed using prcomp function in R on scale-normalized counts. K-means clustering of differentially expressed genes based on fold change values (using cosine distance) was performed using the kmeans function in R.
Functional Classification Based on Gene Ontology, Vitisnet, and Mapman
Grapevine DE genes were subjected to enrichment analyses using: (1) VitisNet annotation within the VESPUCCI grapevine gene expression compendium (http://vespucci.colombos.fmach.it) (Grimplet et al., 2012; Moretto et al., 2016), p-value < 0.01; (2) customized GO annotation and annotated reference, taken from CRIBI annotation (http://www.cribi.unipd.it/), using AgriGO analysis tool (http://bioinfo.cau.edu.cn/agriGO/analysis.php; Du et al., 2010). Enriched GO terms (FDR <0.01) were visualized using the ‘Reduce + Visualize Gene Ontology’ (REViGO) webserver (http://revigo.irb.hr; Supek et al., 2011). Additionally, the differentially expressed genes were visualized in the context of biotic stress pathway using the GrapeGen 12Xv1 annotations version (Lijavetzky et al., 2012) with the help of MapMan tool (Thimm et al., 2004).
Results
Botrytis cinerea Inoculation of Grapevine
Grapevine flowers were inoculated with a GFP-labeled B05.10 strain at full cap-off stage by placing 300 conidia around the receptacle area, and the infection was monitored until ripening, for 12 weeks (Figure 1). No visible symptoms or sign of the fungus growth were observed until full coloring. The proportion of berries carrying B. cinerea at hard-green berry stage (4 wpi; Figure 1B) derived from flower inoculation was checked by plating out on selective media (Figure 2). B. cinerea was present quiescently on 80% of the asymptomatic berries in the samples washed or not with water; however, the proportion dropped to 40% when the berries were surface sterilized, suggesting that the fungus mostly resides in the first few outer epidermal cell layers, those affected by the sterilization procedure. At 10 wpi, at full color change, bunches were bagged with plastic bags to increase humidity and favor B. cinerea growth (Barnes and Shaw, 2002; Elmer and Michailides, 2004; Carisse, 2016). Two weeks later, egression of B. cinerea was observed (Figure 1C), and cross checking the strain using fluorescence microscopy confirmed that the strain was the GFP-labeled B05.10 inoculated at cap-off stage (Figure 1D).
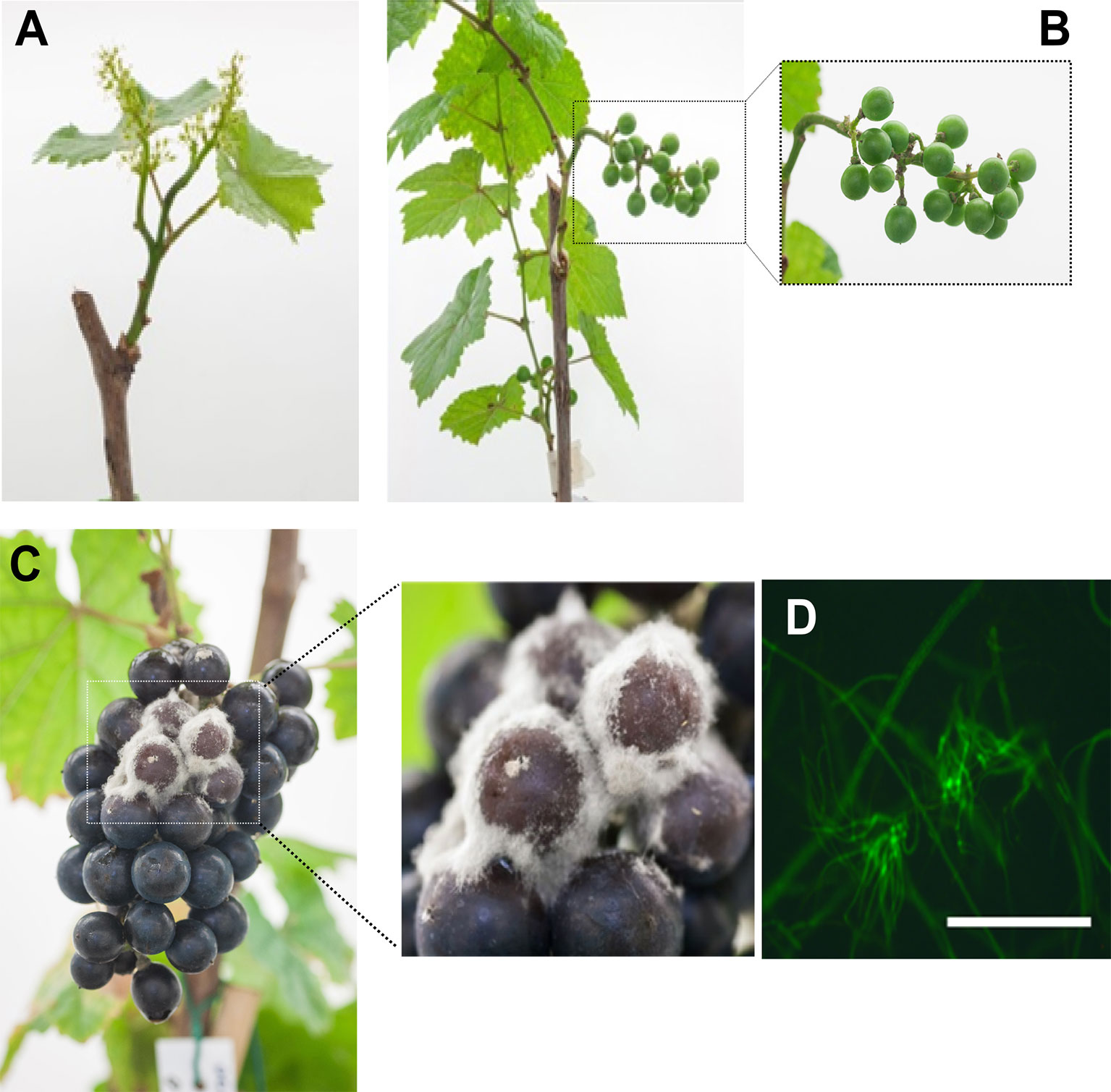
Figure 1 Botrytis cinerea infected grapevine flowers and their development until berry ripening. (A) Flowers 24 hours post inoculation with GFP-labelled B05.10 strain at full cap-fall stage (EL25/26). (B) Healthy looking, asymptomatic, hard-green berries at 4 weeks post inoculation (wpi). (C) Egression of B. cinerea at ripening (12 wpi). (D) Fluorescence of mass of mycelia isolated from the outgrown B. cinerea; white bar represents 50 µm.
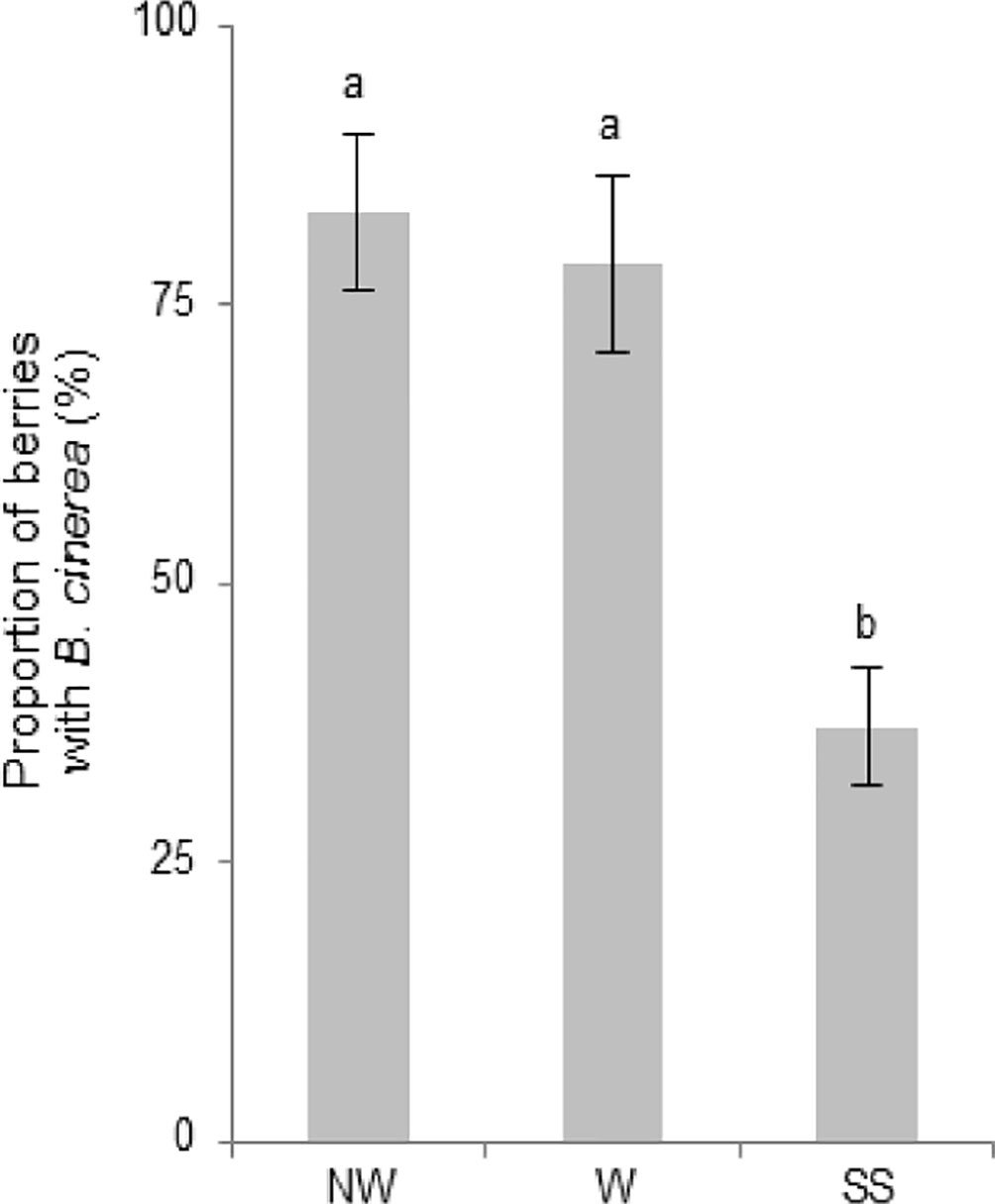
Figure 2 Proportion of berries showing B. cinerea at 4 wpi, following flower inoculation with the fungus, as determined by plating out. Plating out was made on selective media (PDA with Hygromycin B, 70 µg/ml) to check the presence of quiescent B. cinerea before (NW) or after washing (W), or after surface sterilization (SS). Mean proportion of berries (8–10 berries from each of six biological replicates) showing GFP-labeled B05.10 growth on the selective media. Error bars indicate standard error. Mean proportions labeled with the same letter are significantly not different, according to Tukey’s honestly significant difference test (P ≤ 0.05), using one-way ANOVA.
Dual Transcriptomic Analysis of Grape Berries and B. cinerea During Their Interaction
Hard green (4 wpi) and ripe berries (12 wpi), which were mock- and Botrytis-inoculated at cap-off stage, were harvested in three biological replicates for dual (plant and fungus) transcriptome analysis using the RNA-seq method. The fraction of reads uniquely mapped to the Vitis vinifera reference genome ranged from 13 to 88%, the smaller proportion being from samples with egressed B. cinerea. The fraction of reads uniquely mapped to B. cinerea reference genome was below 1% for the 4 wpi and pre-egressed samples, up to 67% for the B. cinerea egressed samples, and up to 78% for PDB grown Botrytis samples (Supplemental Table S2).
The biological variability of all the samples was assessed using PCA on the gene expression data. Concerning grapevine data, samples were largely separated by growth stage along the first principal component, but within each growth stage most of the variation in gene expression was explained by the infection process (Figure 3A). With regard to B. cinerea, the PCA highlighted remarkable difference in gene expression between the egression stage on berries at ripening and the growth in liquid PDB medium (Figure 3B).
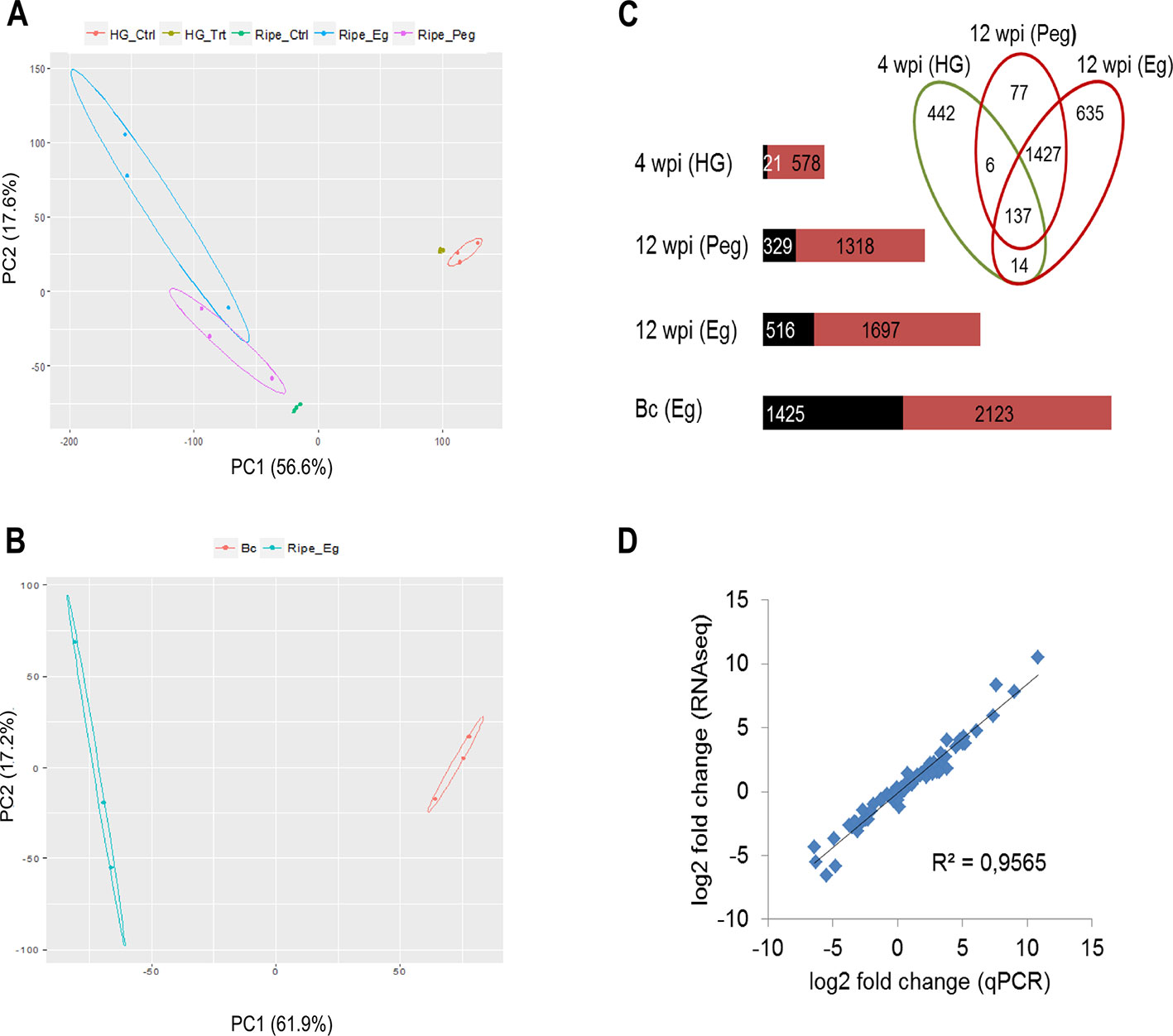
Figure 3 Global evaluation of the RNA-seq data and of the differentially expressed (DE) genes. PCA displaying the biological variations among samples, for grapevine genes (A) and B. cinerea genes (B). Ctrl, mock inoculated; Trt, B. cinerea inoculated; Bc, Botrytis cinerea; HG, hard-green berry; Eg, B. cinerea egression state; Peg, B. cinerea pre-egression state. Raw count data were used after precision weight was calculated by the voom method (Law et al., 2014). (C) Number of DE genes (P < 0.01, absolute fold change > 2.0) upon B. cinerea infection at 4 weeks post inoculation (wpi) and 12 wpi in grape berry and during egression in B. cinerea; upregulated genes (red) and downregulated genes (black). The Venn diagram was generated using Venny v2.1.0 (Oliveros, 2015). Bc, Botrytis cinerea; Eg, egression; HG, hard green; Peg, pre-egession. (D) Correlation of gene expression values obtained by RNA-seq and qPCR. Relative expression levels were calculated for 18 Vitis genes and an R2 value of 0.96 was obtained comparing the results obtained with the two techniques.
For grapevine genes, differential expression was computed between Botrytis- and mock-inoculated berries imposing a p-value < 0.01 and an absolute fold change > 2 (Supplemental Figure S1). At 4 wpi, 599 genes of grapevine were differentially expressed (DE) due to B. cinerea infection, whereas the number increased to 2,296 at 12 wpi (Figure 3C, Supplemental Tables S3 and S4). Only 157 genes were in common, suggesting that the host response is quite different at molecular level at the two growth stages. Concerning B. cinerea genes, for samples at 4 wpi and at 12 wpi (pre-egressed), it was not possible to compute DE genes due to limited amount of fungal RNA in the samples (Supplemental Tables S6 and S7). However, for the egression stage 3,548 DE genes were obtained from the comparison with PDB cultured Botrytis (Figure 3C and Supplemental Table S5). To gain more information from the samples at 4wpi and 12 wpi (pre-egressed), qPCR assay was used to study the expression profile of selected fungal genes.
The gene expression values obtained from RNA-seq were validated using qPCR assay. To this end, the expression of 18 grapevine genes having different expression profile from RNA-seq (Supplemental Table S8) were analyzed and a strong correlation (R2 = 0.96) was observed between the results obtained with the two techniques (Figure 3D).
The total number of DE genes of the grapevine berries (2,738), considering both hard-green and ripe stages, were grouped into 12 distinct clusters according to their expression pattern, which can be grouped in six major expression profiles (Figure 4). Profile A comprised cluster 1 and 2, where almost all of the genes were induced both in hard-green and ripe stages, albeit with various extent. Genes in profile B, combining cluster 3 and 4, exhibited an induced expression trend due to quiescent B. cinerea in the hard-green berry but down-regulated or unaffected during pre-egression and egression stages of the pathogen at ripening. Profile C (cluster 5) included genes whose expression was not affected during B. cinerea quiescence in the hard green berry and pre-egressing at ripe stage, however, they were induced during egression. Genes in profile D (cluster 7, having only 13 genes) were down-regulated at hard-green and not affected at ripe stages. Majority of the genes in profile E were up-regulated during pre-egression and egression at ripening, while the opposite was the case for genes in profile F, where in both profiles the genes were not affected by quiescent B. cinerea in hard-green berry.
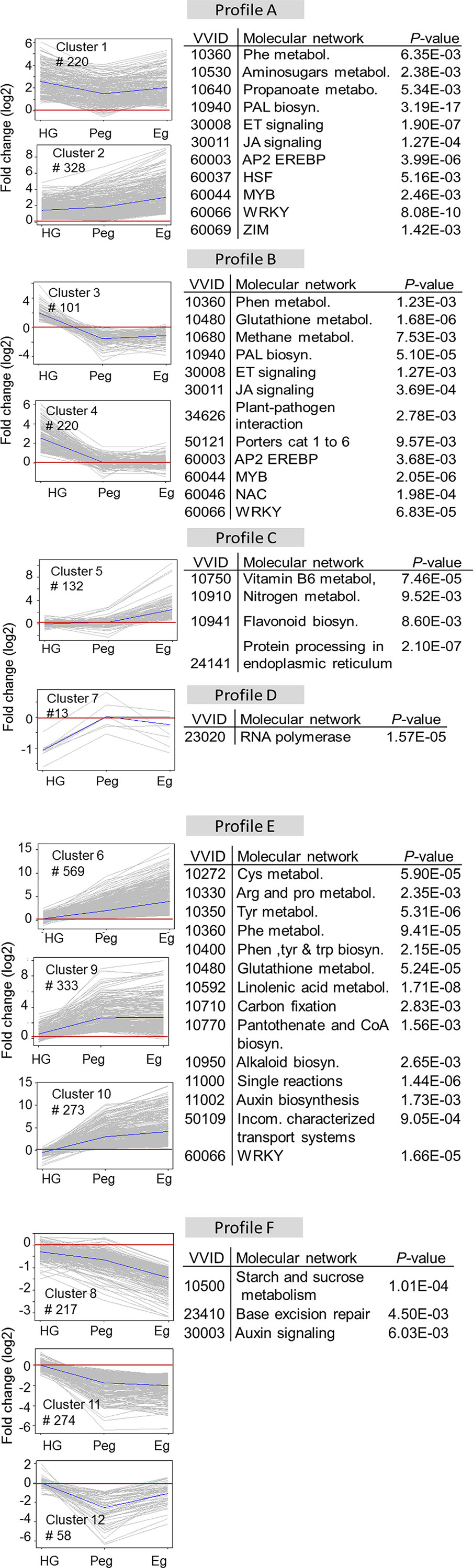
Figure 4 Profiles of grapevine berry trancripts at hard-green and ripe stages in response to B. cinerea inoculation. K-means clustering of grapevine genes based on the cosine distance of their log2 (fold change) values. Genes that showed at least twofold expression difference with P-value < 0.01 were considered, and clustered into 12 clusters. The clusters were grouped into six major profiles (A, B, C, D, E, and F). Molecular enrichment analysis based on VitisNet is provided for each group.
The molecular network enrichment analysis of the gene set of each profile, based on VitisNet annotation, showed an abundance of transcripts in functional classes which are usually affected by biotic stress (Figure 4). A considerable number of genes annotated as belonging to phenylpropanoid biosynthesis, phytohormone signaling, and encoding transcriptional factors (TF) were mainly represented in profile A and B; genes involved in protein processing in profile C; genes of amino acid and glutathione metabolisms in profile E; and genes of carbohydrate metabolism in profile F.
Transcriptional Alterations of B. cinerea During Quiescent Infection, at 4 wpi
The number of reads mapping on the B. cinerea transcriptome detected in the inoculated samples at 4 wpi was very low. The reason could largely be linked to a reduced fungal biological activity as well as to the little fungal mass present at the quiescent stage. Only about 20% of the B. cinerea genes (1,926) had at least one raw read in all of the three biological replicates. Within this set, those represented by an average of at least ten reads (only 289) were considered as in planta expressed fungal transcript and they were functionally annotated using Blast2GO (Conesa et al., 2005) and Amselem et al. (2011) (Supplemental Table S6). Using the Combined Graph Function of Blast2GO, the GO slim terms metabolic processes, structural constituent of ribosome, and intracellular were mostly represented in the 289 genes (Supplemental Table S9). Fifteen genes from this group, selected on the basis of their function, are presented in Table 1. The expression profile of nine of them, involved in functions such as cell wall metabolism, redox-reaction, and transcriptional regulation, was further examined using qPCR assay (Figure 5). As depicted in Figure 5, all with the exception of Bcin07g01540 and Bcin13g05810 had a higher relative expression during quiescent infection at hard green stage than during initial infection, pre-egression, and egression stages.
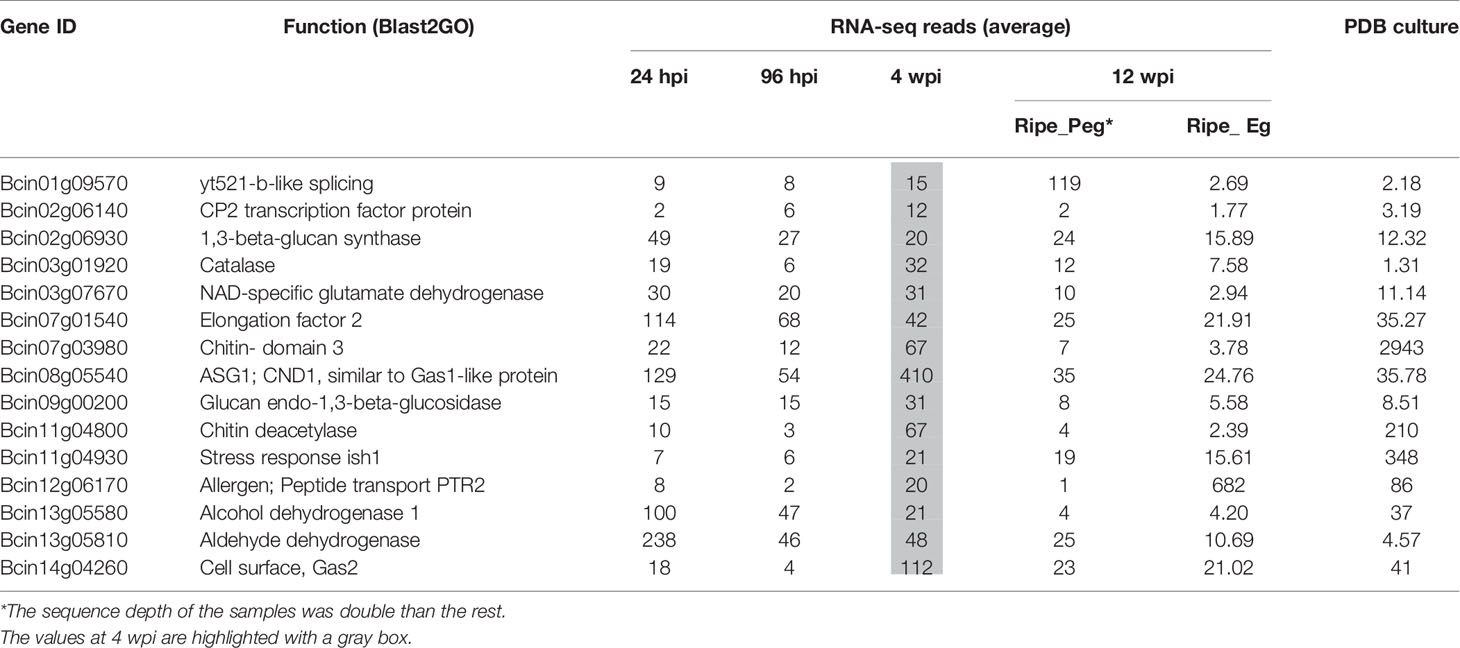
Table 1 Selected B. cinerea genes expressed in planta in the hard-green berry (4 wpi), with different functions, with their raw RNA-seq reads.
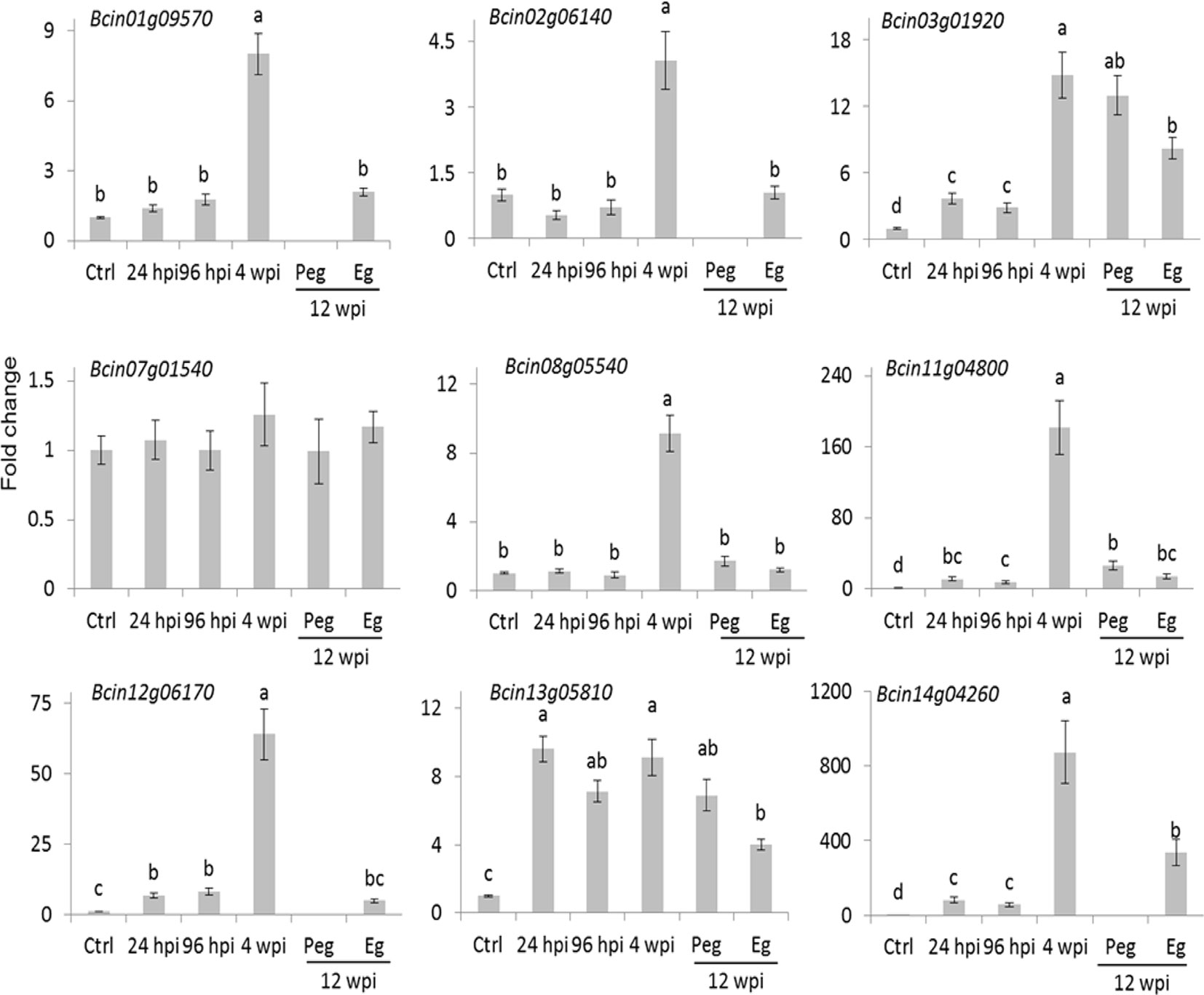
Figure 5 Expression profile of selected B. cinerea genes having higher raw reads at hard-green berry stage relative to PDB-cultured B. cinerea expression. Bars represent fold change of samples at 24 and 96 hours post inoculation (hpi; flower infection), 4 weeks post inoculation (wpi; quiescent infection at hard-green berry stage), and 12 wpi (pre-egression, Peg, and egression, Eg, stages of B. cinerea at ripening) relative to the PDB-cultured B. cinerea (Ctrl) expression. Normalization based on the expression levels of ribosomal protein L5, BcRPL5, and α tubulin, BcTUBA, was carried out before calculating fold changes. Error bar represents standard error of the mean of three biological replicates. Expression values followed by a common letter are significantly not different between samples, according to Tukey’s honestly significant difference test (P ≤ 0.05), using one-way ANOVA of log2(NRQ).
These results suggested that the biological activities of the fungus were not totally switched off during quiescent infection. The expression of Bcin01g09570 and Bcin07g01540 genes, which encode putative yt521-b-like splicing and elongation factor 2 proteins, respectively, and the numerous genes encoding for ribosomal proteins (Table 1 and Supplemental Table S6) indicated instead that protein synthesis activities were carried out during quiescent infection stage. Moreover, the expression of stress and defense related genes such as Bcin12g06170, encoding a protein similar to an allergen, and Bcin11g04800, encoding a putative chitin deacetylase protein, highlighted that the interaction between the pathogen and the plant was not passive. Chitin deacetylase activity has been speculated to be involved in protecting the fungal cell wall from degradation by plant chitinases (Deising and Siegrist, 1995; El Gueddari et al., 2002). Moreover, Bcin08g05540, encoding putative CND1 protein, Bcin14g04260, annotated as a putative cell surface protein and Gas2, and Bcin02g06140, encoding a putative CP2 transcription factor protein, appeared to be expressed more during quiescent infection at 4 wpi as compared to initial and egression stages of infection (Figure 5). These proteins should be involved in maintaining cell wall integrity (Garrett-Engele et al., 1995; Paré et al., 2012).
In addition to cell wall remodeling in which the fungus was engaged, the involvement of genes detoxifying alcohols, aldehydes, and ROS such as Bcin13g05810, Bcin13g05580, and Bcin03g01920 (Table 1 and Supplemental Table S6) during quiescent state indicated that the fungal cell was trying to overcome stresses. In fungal cells, it is known that stress causes ROS production that can lead to aldehydes and alcohols accumulation (Asiimwe et al., 2012).
In summary, the extremely low number of B. cinerea genes expressed in planta seconded by the absence of any macroscopically noticeable disease progress at 4 wpi suggests that the pathogen was at its basal metabolic activity, but some specific stress related functions were in place.
Response of Hard-Green Berries to Quiescent B. cinerea, at 4 wpi
In contrast to the fungus, for which not many transcripts were observed, 599 grapevine genes were regulated due to the quiescent presence of B. cinerea, only 21 genes of them being down-regulated (Figure 3C and Supplemental Table S3). In this set of Botrytis-induced genes, functional classes related to responses to stress, amino acid metabolism for redox activity and phenylpropanoid pathways, signaling, and TFs were over-represented (Table 2 and Supplemental Table S10). The visualization of individual gene responses in biotic stress pathway using MapMan tool also confirmed a remarkable induction of genes related to signaling, TFs, proteolysis, PR proteins, and secondary metabolism (Figure 6).
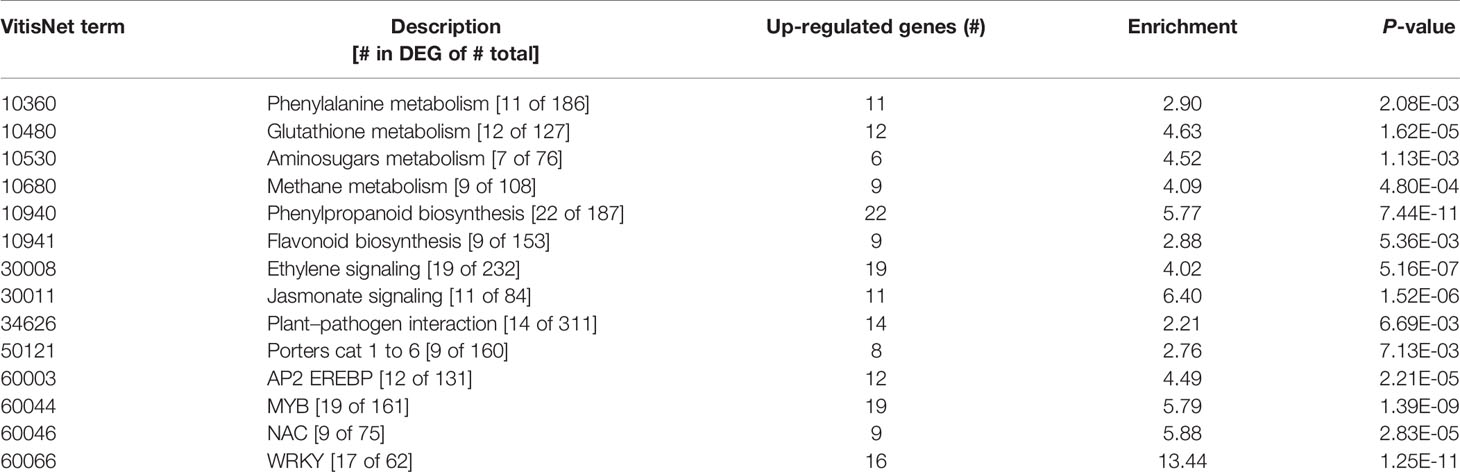
Table 2 Functional classes enriched in the differentially expressed genes at hard-green berry stage due to quiescent B. cinerea, using VitisNet.
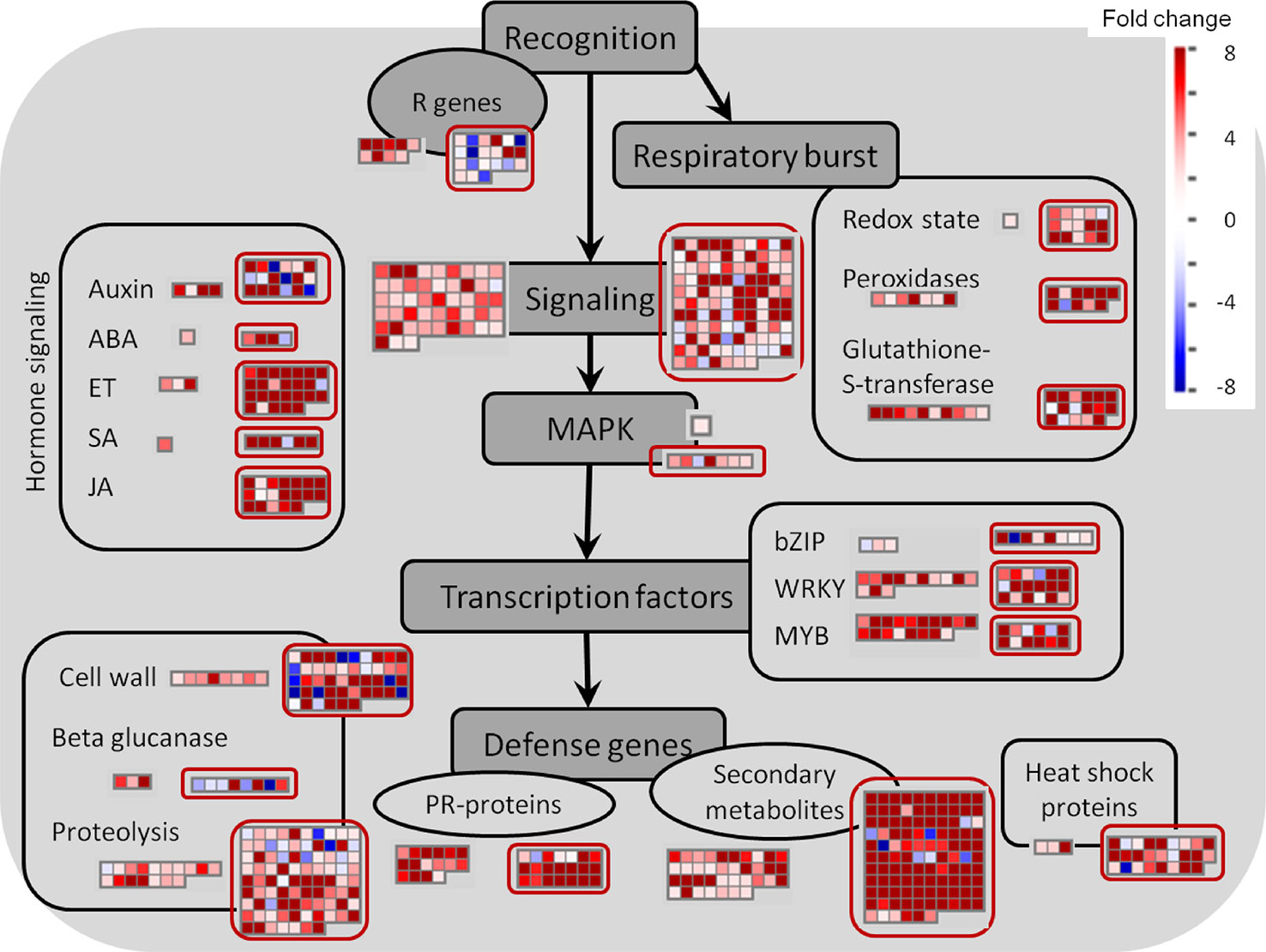
Figure 6 Biotic stress overview of hard-green and ripe berries (red circles) due to quiescent and egressed B. cinerea infection, respectively, as visualized by MapMan. Up-regulated and down-regulated genes are shown in red and blue, respectively. The scale bar displays fold change values. ABA, abscisic acid; ET, ethylene; JA, jasmonic acid; MAPK, mitogen-activated protein kinase; SA, salicylic acid.
About one-sixth of the differentially expressed genes were those involved in signaling, dominated by the receptor-like protein kinases (RLKs) (Table 3 and Supplemental Table S3). Of the membrane-localized RLKs which were switched on during infection initiation at flowering, Clavata1 receptor kinase (CLV1), Brassinosteroid insensitive 1-associated kinase 1 (BAK1), and Wall-associated kinase 1 (WAK1) (Haile et al., 2017) were found induced in the hard-green berry as well during quiescent infection. However, the involvement of WAK1, a damage associated pattern receptor which recognizes plant cell wall–derived oligogalacturonides due to cell wall degradation (Brutus et al., 2010), during quiescent interaction was not obvious. From the transcriptional alteration of RLKs, lectin protein kinase and protein kinase 1 seem to be quiescent-stage-specific. The putative orthologues in other species are associated to plant immunity (Guidarelli et al., 2014), in particular the latter is known to mediate signaling in response to B. cinerea (Abuqamar et al., 2008). In addition, transcripts related to Ca2+ mediated signaling (such as calcium- and calmodulin-binding proteins) and oxidative stresses (mainly GST and cytochrome P450 monooxygenases) were also found to be possibly involved in the ongoing berry immunity.
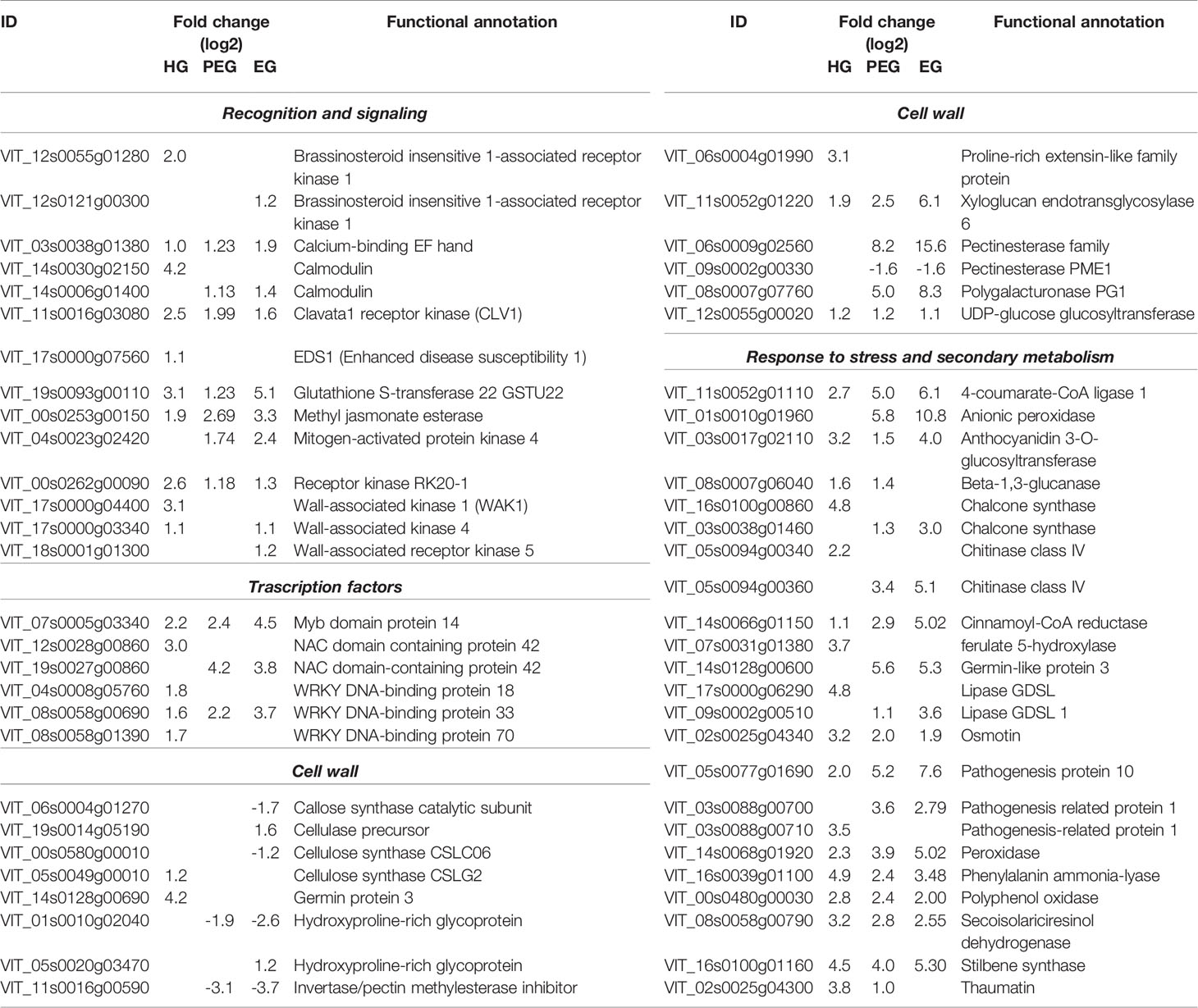
Table 3 Selected differentially expressed grapevine genes due to B. cinerea infection (at 4 and 12 wpi).
The quiescent B. cinerea prompted the expression of key TFs playing an important role in plant–microbe interaction (Table 3 and Supplemental Table S3). Most prominent was the MYB TF family: 21 genes encoding 14 different MYB proteins, including VvMYB14 and VvMYBPA1 which respectively regulate stilbene and proanthocyanidin biosynthesis. VvWRKY33, known to regulate defense response against pathogens (Birkenbihl et al., 2012; Merz et al., 2015), was also among the TFs involved in the hard-green berry response. From the NAC and WRKY TF families, it is worth mentioning the ortholog to NAC042, regulates phytoalexin biosynthesis (Saga et al., 2012); the ortholog to WRKY51, mediates the repression of JA signaling in a SA- and low-oleic-acid-dependent manner (Gao et al., 2011); and the orthologs to NAC036, WRKY18 and WRKY70, which regulate SA biosynthesis and SA signal transduction (Wang et al., 2006) in Arabidopsis. The up-regulation of SA signaling marker genes, such as PR1 and EDS1, is a further indication of SA involvement in enhancing the defense ability of the hard-green berry. In addition, the induction of 3 ACC oxidase and 12 AP2/ERF genes underlines that ET signaling was also in place during the interaction at quiescent state. AP2/ERF factors indeed play a key role in plant–pathogen interactions (Gutterson and Reuber, 2004; Licausi et al., 2013). Genes related to auxin metabolism were also modulated.
Intriguingly, differential regulation was also observed for genes encoding various PR proteins and enzymes of the phenylpropanoid pathway and of the flavonoid and stilbenoid biosynthesis during quiescent infection (Table 3 and Supplemental Table S3). For some of these genes, the induction was nearly 50 fold. PR proteins and its regulator encoding genes included: VvPR10s and VvWRKY33, previously reported as involved in the defense against B. cinerea and Plasmopara viticola in grapevine (Merz et al., 2015; Haile et al., 2017); VvPRs encoding β -1,3-glucanase, different classes of chitinases, osmotin, thaumatin, and cystatin, known to interfere with B. cinerea growth (Pernas et al, 1999; Monteiro et al., 2003), .
Considering polyphenol biosynthesis, genes involved in monolignol, flavonoid, and stilbenoid biosynthesis pathways, such as phenylalanine ammonia-lyase, stilbene synthase, anthocyanidin 3-O-glucosyltransferase, chalcone synthase, and cinnamoyl-CoA reductase were found differentially regulated in the hard-green berry with quiescent Botrytis (Figure 7 and Supplemental Table S3). A number of compounds of the phenylpropanoid pathway were also quantified by UHPLC-DAD-MS (Figure 7 and Supplemental Table S11). The strongest effect was recorded on flavonoids and stilbenoids, in particular on compounds known to mediate defense against pathogens: resveratrol, viniferins, ampelopsin, miyabenols, isohopeaphenol, catechin, and proanthocyanidins (Jersch et al., 1989; Goetz et al., 1999; Pezet et al., 2003a; Favaron et al., 2007; Hammerbacher et al., 2011). For the stilbenoid class, most of the compounds were below the detection limit in control samples.
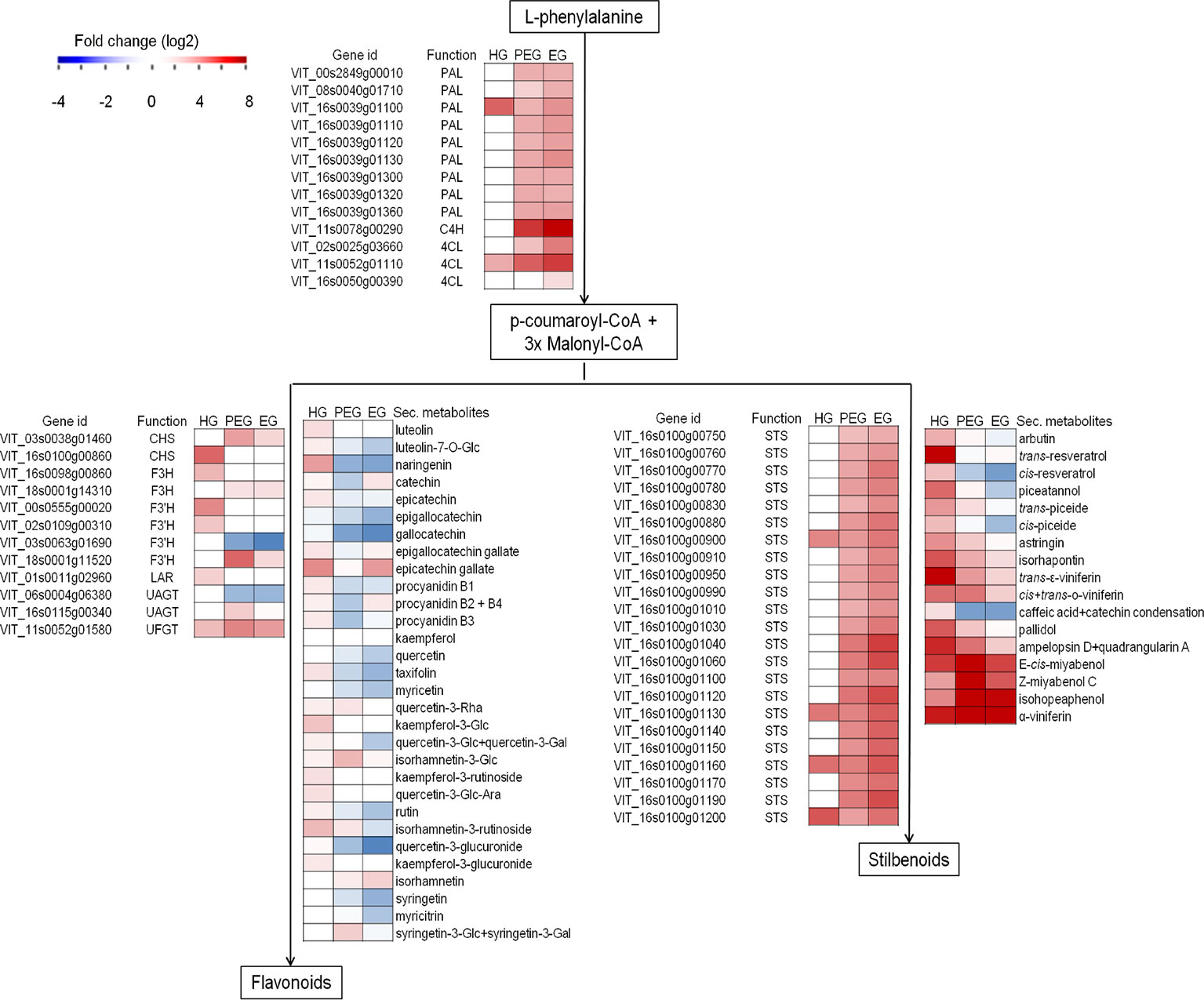
Figure 7 Secondary metabolism-associated genes and metabolites modulation in grapevine berries in response to B. cinerea. Heatmap of gene expression (from RNA-Seq result) and secondary metabolite concentration (µg/g fw, from HPLC-DAAD-MS) expressed as fold change. Fold change for secondary metabolites was computed based on the ratio of average values in B. cinerea- and mock-inoculated berries. HG, hard-green berry; PEG, ripe berry with pre-egressed B. cinerea; EG, ripe berry with egressed B. cinerea. Central phenylpropanoid biosynthetic pathway: PAL, phenylalanine ammonia-lyase; C4H, cinnamate 4-hydroxylase; 4CL, 4-coumarate-CoA ligase. Flavonoid biosynthetic pathway: CHS, chalcone synthase; F3H, flavonone-3-hydroxylase; F3'H, flavonoid-3'-hydroxylase; LAR, leucoanthocyanidin reductase; UAGT, UDP-glucose:anthocyanidin 5,3-O-glucosyltransferase; UFGT, UDP-glucose:flavonoid 3-O-glucosyltransferase. Stilbenoid biodynthesis pathway: STS, stilbene synthase.
Taken together, even though we did not observe any known virulence related genes of the pathogen which can provoke response in the hard-green berries, it seemed, that the berries were alerted with enhanced immunity as it recognizes non-self organism, which in turn helped the berry to contain the pathogen. This gives an interesting insight that there is a molecular communication going on between the quiescent B. cinerea and the hard-green berry.
Transcriptional Alterations of B. cinerea During Pre-Egression and Egression Stages, at Ripening
At ripening (12 wpi), two kinds of berries were collected from Botryits-treated samples: berries without visible B. cinerea outgrowth (pre-egressed B. cinerea) and berries with visible signs of B. cinerea outgrowth (egressed B. cinerea). Both samples were subjected to RNA-seq analysis. In the samples with pre-egressed B. cinerea, as in hard-green berry samples, the number of fungal transcripts was unfortunately very low, even though the sequencing depth was doubled. Moreover, in one of the biological replicates (replicate 2) the growth of the fungus was more advanced than in the rest two, as inferred from the number of fungal transcripts obtained (Supplemental Table S2). Taking the same threshold used in the hard-green berry samples (an average of at least ten reads) in the two biological replicates, excluding the replicate two, 431 genes were selected for functional annotation and further analysis (Supplemental Table S7). The most represented functional classes in this gene set are provided in Supplemental Table S12.
Within the 431 genes, there were several genes encoding proteins functionally associated to the infection process, among which dyp-type peroxidase and galactose oxidase, involved in generation and detoxification of ROS (Schumacher et al., 2015); polygalacturonase, deployed in pectin degradation; glyoxal oxidase and oxalate decarboxylase, both catalyzing oxalate; different types of oxidoreductases; and cerato-platanin BcSPL1, a small protein required for full virulence (Frías et al., 2011; Frías et al., 2014). Interestingly, these genes showed a low number of raw reads or were not expressed at all at 4 wpi (Table 4), indicating that the fungus was in a different physiological state in the ripe berries before egression as compared to that in the hard-green berries, though there was no any apparent disease symptom in both cases.
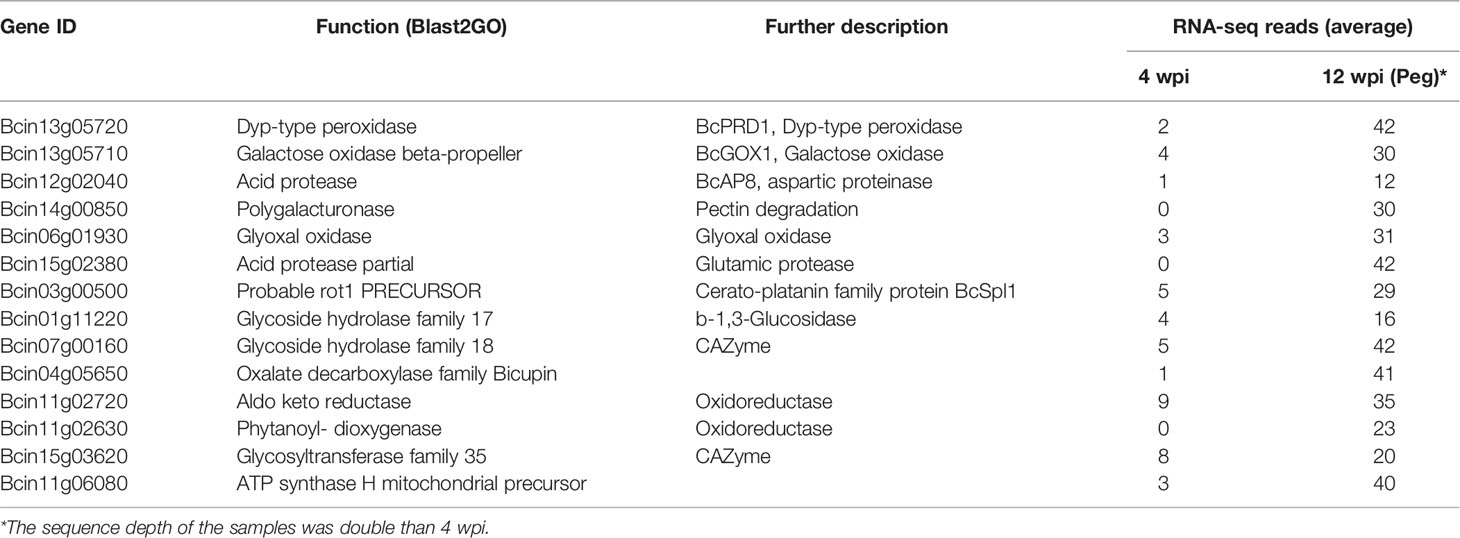
Table 4 Selected B. cinerea genes having more RNA-seq reads at pre-egression (12 wpi, on ripe berry) than at quiescence (4 wpi, on hard-green berry).
Unlike, in the egressed samples, 86% of total B. cinerea transcriptome was expressed, about 10,000 of the total 11,701 predicted genes (van Kan et al., 2016). Such a massive transcriptional activity was not seen in the other infection stages, possibly due to the low amount of the fungus, but likely also to a reduced transcriptional activity at those stages. In other words, at ripening, the time, the status of the host tissue, and the environmental conditions, components of the disease pyramid, were conducive for B. cinerea to egress and grow vigorously, as observed in Figure 1C.
Compared to the transcriptional changes of B. cinerea cultured in PDB, 3,548 genes were differentially regulated during egression at ripening (Figure 3C and Supplemental Table S5). These DE genes are over-represented in metabolic processes, ion binding, catalytic and oxidoreductase activities, cytoplasm, intracellular part functional classes (Supplemental Table S13). Genes encoding carbohydrate-active enzymes and others involved in plant cell wall degradation (Espino et al., 2010; Blanco-Ulate et al., 2014), such as Bcin10g06130 and Bcin14g01630, encoding pectinases, Bcin03g01680, encoding a polygalacturonase, and Bcin07g06480 and Bcin15g03080, encoding cutinases, were expressed more during egression than in PDB medium. Other virulence and/or growth related genes having similar expression trend as those mentioned above were: ROS producers and scavengers like Bcin03g03390, Bcin13g05710, and Bcin13g05720 (Rolke et al., 2004; Schumacher et al., 2015); characterized aspartic proteases Bcin12g02040 and Bcin12g00180 (ten Have et al., 2010); membrane transporters, mostly the ATP-binding cassette; and the botcinic acid and botrydial phytotoxins Bcin12g06390 and Bcin12g06380 (Siewers et al., 2005; Dalmais et al., 2011). On the other hand, known virulence genes like BcPG1 (Bcin14g00850), BcGST1 (Bcin10g00740), BcBOA6 (Bcin01g00060), and BcSPL1 (Bcin03g00500) had similar or lower expression level during egression as compared to PDB cultured Botrytis. This, however, does not mean that they have not played any role in the necrotrophic stage of infection during egression at ripening, as the number of reads of these genes was reasonably high at this stage (Table 5).

Table 5 RNA-seq reads of key B. cinerea virulence genes which were not considered as differentially expressed in comparison to PDB culture of B. cinerea.
Response of Ripe Berries to B. cinerea, at Pre-Egression and Egression States
Grapevine berries responded to the necrotrophic colonization of the fungus, during egression, by reprogramming the transcription of 2,213 genes (Figure 3C and Supplemental Table S4). Of these genes, 1,564 were already differentially regulated in pre-egressed samples. The GO enrichment analysis performed on the DE genes of pre-egressed and egressed samples showed a high overlap in the enriched biological processes: secondary metabolic process, response to stimulus, catabolic process, and transport were among the shared enriched functional classes by both samples (Supplemental Table S14).
Besides the very high overlap of DE genes of the pre-egressed and egressed samples, there existed also a general similarity in the expression trend (up- or down-regulation) of these common genes shared by both samples (Figure 3C and Supplemental Table S4). As a result, we used the DE genes of the egressed stage to visualize the biotic stress pathway (Figure 6), via MapMan, as it can give enough insight of the pre-egressed stage as well. Figure 6 showed that the transcriptional changes involved in biotic stress were huge. Although it might seem surprising, it is not uncommon to see such an extensive modulation of defense related genes in infected plant tissues also when the pathogen invades the plant host tissues (for example in: Alkan et al., 2014; Agudelo-Romero et al., 2015; Kelloniemi et al., 2015). This could be due to the futile attempts of the infected tissue reacting against the pathogen and/or to the transcriptome attributes of other cell layers, not yet colonized, as it is very difficult to spatially resolute the tissues where the pathogen is growing from those which are not yet colonized. The comparison of the two samples highlights the presence of several genes associated to auxin, JA and ET signaling and cell wall modification uniquely modulated in the ripe berry in response to B. cinerea (Figure 6).
Hormones interactions during plant defense are extremely complex. At transcriptional level, it appeared that auxin, ET, JA, and SA were involved (Supplemental Table S4). However, the co-activation of SA and JA pathways is uncommon and very interesting (Oirdi et al., 2011; Kelloniemi et al., 2015) and therefore should be further investigated. Genes encoding PR1, a SA marker, and ZIM-domain1, a JA marker, were all induced (Table 3 and Supplemental Table S4).
Polyphenol biosynthesis pathway was also affected by the egression of B. cinerea at ripening (Table 3 and Supplemental Table S4), though the transcription of genes encoding dihydroflavonol-4-reductase and flavanone 3-hydroxylase, both involved in flavonoid biosynthesis, was not affected. In line with this evidence, almost all of the quantified flavonoids were significantly lower or not significantly different in the berries with egressed B. cinerea as compared to the control (Figure 7 and Supplemental Table S15). On the other hand, genes encoding other key enzymes such as cinnamate 4-hydroxylase, 4-coumarate-CoA ligase 1, and stilbene synthase which are involved in the biosynthesis pathway were induced. With regard to stilbenoid content, the concentration of piceide, miyabenol, viniferins, and isohopeaphenol was high in berries with pre-egressed Botrytis, intermediate in berries with egressed Botrytis, and very low in healthy berries (Figure 7 and Supplemental Table S15).
Most of the genes involved in monolignol biosynthesis (such as cinnamoyl-CoA reductase and cinnamyl alcohol dehydrogenase) were differentially regulated (Supplemental Table S4), unlike a few but important genes like caffeoyl-CoA O-methyltransferase (CCoAMT) and ferulate 5-hydroxylase (F5H) encoding ones, which were not differentially regulated. CCoAMT is involved in ferulic esterification and lignification process in response to pathogen attack in grapevine (Busam et al., 1997). From the polyphenol analysis, caffeate and ferulate, substrate for CCoAMT were not detected in any of the samples at ripening (Supplemental Table S15). This perhaps might suggest that lignification is not part of plant defense component during ripening. Rather, a number of genes encoding proteins involved in ripening-associated cell wall extensibility and disassembly like xyloglucan endotransglucosylase and polygalacturonase and pectinesterases (Nunan et al., 2001; Deytieux-Belleau et al., 2008) were highly upregulated in ripe berries with pre-egressed and egressed Botrytis (Supplemental Table S4). Pectin methylesterases (PMEs) are suggested to be involved both in cell wall loosening and strengthening (Micheli, 2001), in particular PMEs are involved in the de-esterification in muro of pectin which becomes more susceptible to the degradation by pectic enzymes secreted by B. cinerea during the initial stages of infection (Lionetti et al., 2012). However it seemed here that cell wall loosening prevailed over cell strengthening as there were a lot of induced polygalacturonases which degrade polygalacturonans, made accessible by the pectinesterases.
Discussion
In a previous study, grapevine flowers were challenged by placing suspension of B. cinerea conidia to induce infection (Haile et al., 2017). The results showed that fungal genes encoding virulence factors and proteins known to contribute to the infection program were highly induced. Consequently, the flower reprogrammed its transcriptome which resulted in increased expression levels of genes involved in reduction–oxidation processes, genes encoding antimicrobial proteins, and genes of the polyphenol biosynthesis pathway, for the production of phytoalexins and precursors for cell wall toughening (Haile et al., 2017). These defense reactions of the flower appeared to be able to put B. cinerea into quiescence. To know more of the later stages of the infection process, this study was conducted with an in-depth look at the molecular communication between the fungus and the berry at hard-green (4 wpi) and at ripe (12 wpi) stages. Figure 8 summarizes the most important events occurring from infection initiation (24 hpi) till fungal egression (12 wpi) passing through entering in quiescence (96 hpi) and quiescence (4 wpi) phases occurring both in the grapevine and in the fungal cells, respectively.
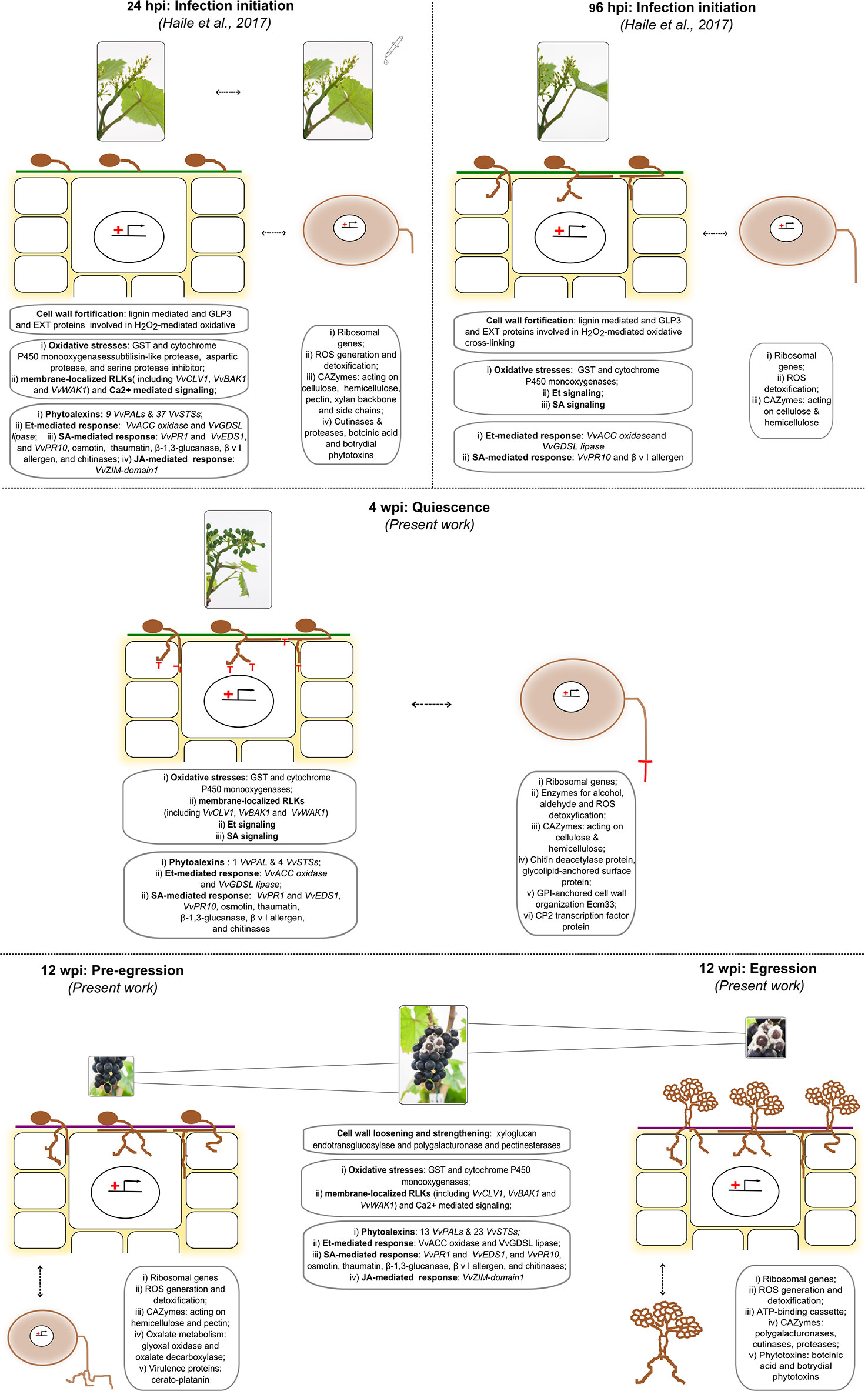
Figure 8 Depiction of the most important events from infection initiation (24 hpi) till fungal egression (12 wpi) passing through entering quiescence (96 hpi) and quiescence (4 wpi) phases occurring both in the grapevine and in the fungal cells, respectively. The events at the initial stages (24 and 96 hpi) are taken from Haile et al. (2017), while the other came out from the present work.
In the very early stage of quiescence in the flowers, ribosomal genes were prevalent in the in planta expressed B. cinerea genes (Haile et al., 2017). A similar high proportion of ribosomal genes were also observed within the in planta expressed genes of B. cinerea in the hard-green berry at 4 wpi (Supplemental Table S6). Nevertheless, there was no known virulence-related gene in the Botrytis genes expressed in planta at 4 wpi. Yet, other biological activities helping the fungus to stay “alive” were likely on-going since elongation factors, ATP synthesis, and ATP-dependent molecular functions related genes were transcriptionally active. For example, Bcin15g02120 (glyceraldehyde-3-phosphate dehydrogenase) and Bcin16g04800 (malate dehydrogenase) genes involved in glycolysis and tricarboxylic acid cycle for energy metabolism were expressed. Also among the expressed genes were an ATP-dependent cell division cycle protein 48 (p97/valosin-containing protein, Bcin08g03700) gene, involved in cell cycle and transcriptional regulation (Wang et al., 2004), and a number of ATP-dependent membrane transporter genes. As to the source of energy, the quiescent B. cinerea expressed 34 CAZyme genes suggesting that also in this phase it is still capable of extracting energy from the host.
The absence of detectable pathogenic progression of the quiescent B. cinerea is possibly due to the continued induction at 4 wpi of those transcripts that were found involved in plant defense at bloom at initial infection stage (Haile et al., 2017). Such continued activation of defense pathways between appressoria formation and quiescent stages of infection was also reported in unripe green tomato infection by Colletotrichum gloeosporioides (Alkan et al., 2014). The expressions of VvWRKY33 gene, also correlates with the expression of VvPR10 genes in response to defense in grapevine (Merz et al., 2015; Haile et al., 2017), and genes of different families of PR proteins, including PR10, were highly induced in the hard-green berry due to the quiescently present B. cinerea (Supplemental Table S3). The grapevine WRKY33 functional homologue, AtWRKY33, has been shown to be involved in response to biotic and abiotic stresses (Zheng et al., 2006; Jiang and Deyholos, 2009; Li et al., 2011; Birkenbihl et al., 2012). On the other hand, five GDSL lipase encoding genes, whose expression was not affected during flower infection, were strongly upregulated (up to 25-fold) during quiescent infection. No previous report associated these lipases with a defense response against pathogens; however, other lipases, such as GDSL Lipase 1 (Oh et al., 2005), are involved in defense against Alternaria brassicicola and B. cinerea in Arabidopsis in an ET dependent manner regulated by WRKY33 (Oh et al., 2005; Kwon et al., 2009; Birkenbihl et al., 2012).
The induction of PR proteins in the hard-green berry might compel cell wall remodeling in the quiescent B. cinerea. The qPCR assay confirmed that Bcin11g04800, a gene encoding chitin deacetylase, was highly induced during the quiescent phase (Figure 5). Chitin deacetylation is a mechanism used by plant pathogens as well as endophytic fungi to protect their cell wall from being attacked by plant chitinases (Deising and Siegrist, 1995; El Gueddari et al., 2002). Chitin depolymerization into deacetylated chitosan oligomers avoid the binding by plant receptors and the consequential plant immune responses (Petutschnig et al., 2010; Liu et al., 2012). Recently, Cord-Landwehr and colleagues (Cord-Landwehr et al., 2016) demonstrated that chitosan oligomers, deacetylated chitin extracted from an endophytic fungus Pestalotiopsis sp., were not able to elicit plant immunity in rice cell suspension culture. Thus, the enzyme might play an important role, particularly during quiescent phase, to impair the recognition of the quiescent B. cinerea by the plant immunity system. In addition to chitin deacetylase, other genes encoding glycolipid-anchored surface protein and GPI-anchored cell wall organization ECM33, which in yeast are linked to cell wall integrity to ensure viability (Pardo et al., 2004), were also expressed during the quiescent phase, suggesting that the fungus is also actively defending itself besides the basal metabolic activity.
The activation of stilbenoid and flavonoid biosynthetic pathways by grapevine in response to active pathogenic infection is well documented (Langcake, 1981; Jeandet et al., 1995; Keller et al., 2003; Malacarne et al., 2011; Agudelo-Romero et al., 2015; Kelloniemi et al., 2015; Haile et al., 2017). Here we observed that genes encoding essential enzymes of the pathways, such as stilbene synthase, chalcone synthase, flavanone 3-hydroxylase, and anthocyanidin 3-O-glucosyltransferase, were actively engaged during quiescent infection. A key transcription factor regulating stilbene biosynthesis VvMYB14 (Höll et al., 2013) was also modulated. As expected, from the transcriptional analysis results, several polyphenols were also at higher concentration in the inoculated samples. The content of resveratrol and its monomeric (for example astringin, isorhapontin, and piceide) and oligomeric (for example miyabenol, isohopeaphenol, and viniferin) derivatives, known defense compounds Pezet et al., 2003a; Favaron et al., 2007; Hammerbacher et al., 2011), was higher in hard-green berry with quiescent Botrytis than in the control samples.
Relevant transcripts for the synthesis of monolignol precursors (VvPAL, VvCOMT, VvCCoAMT), which increase penetration resistance in the plant cells (Bhuiyan et al., 2009), and other lignin forming enzymes like GLP3 and EXT, were also induced in the hard-green berry. Lignification at the penetration site is one of the major defense mechanisms that plants adopt to stop B. cinerea progress (Cantu et al., 2009; Kelloniemi et al., 2015; Haile et al., 2017). It is interesting to observe the pathway being active at 4 wpi in the hard-green berry.
However, egression of B. cinerea was observed after bunches were bagged for 2 weeks, starting at full coloring (approximately 10 wpi), to create high humidity around the bunch (Figure 1C). At the very start of the egression process, an outgrowth of B. cinerea (or egression) was observed on about 40% of the berries.
During egression about 86% of the B. cinerea transcriptome was expressed, and it encompassed genes functionally annotated as ROS producers and scavengers, CWDE, proteases, and enzymes involved in the synthesis of phytotoxic secondary metabolites, which are sustaining B. cinerea pathogenicity (as reviewed in Nakajima and Akutsu, 2014). Botrytis transcripts belonging to these functional classes were also shown to be involved in the successful infection of ripe grapevine berries and other hosts (De Cremer et al., 2013; Smith et al., 2014; Kelloniemi et al., 2015). However, the expression level of BcPG6 and BcPEL-like1, pectinases which were extremely induced during initial infection at bloom (Haile et al., 2017), was much less both at ripe and in PDB culture. In general, as expected, the transcriptional activity of the pathogen was high during egression. An important question is what signals and/or environmental changes made the quiescent B. cinerea egressing. Although still speculative, it is likely that ripening associated signals and physical and chemical changes play an important role in triggering the transition from the prolonged quiescent to the egression phase.
We have noticed that a lot of grapevine genes involved in cell wall disassembly were induced. Cell wall loosening, cuticular changes, conversion of acids into sugars, and a steadily diminishing of antifungal compounds are reported to favor pathogen egression (Prusky, 1996; Prusky et al., 2013). It has also been shown that the protective role of berry cuticle on B. cinerea infection decreased with ripening (Commenil et al., 1997; Mlikota-Gabler et al., 2003). The structural changes in the cell wall polysaccharides that lead to fruit softening could cause susceptibility to necrotrophic pathogens at fruit ripening (Cantu et al., 2008). These authors showed that suppression of the ripening-associated cell wall loosening genes reduced the susceptibility of ripe tomato to B. cinerea (Cantu et al., 2008). Besides cell wall loosening, sugars and organic acids exudates appearing on ripe berry surface have also the potential to stimulate and promote B. cinerea outgrowth (Padgett and Morrison, 1990; Pezet et al., 2003b; Kretschmer et al., 2007). Furthermore, a shift in plant-hormone synthesis and signaling balance happening during ripening also trigger fungal pathogenicity factors (Prusky, 1996; Prusky et al., 2013).
Considering the transcriptional alterations underwent in the ripe berry, as a response to Botrytis egression, a wide array of defense responses were noticed, suggesting that the tissue under colonization “never gives up” rearranging its defense mechanisms. With regard to stilbenic compounds, surprisingly oligomerization was apparently driven by the presence of the fungus. Oligomerization, according to Pezet et al. (2003a) and Malacarne et al. (2011), increases toxicity. The amount of oligomeric stilbenoids was very little in the control berries and higher in the treated berries especially in the pre-egressed ones. One possible explanation is the fact that the B. cinerea LACCASE 2 enzyme (encoded by Bcin14g02510) which oxidizes resveratrol (Schouten et al., 2002) was extremely induced (128 fold) in the egressed Botrytis.
Last, the evolution of the berry skin tissue is an important component of the berry-Botrytis interaction. It has been noticed that the extent of the expression of cell wall modifying genes increases toward maturity. It is actually a phenological cue that once the seeds are matured, cell wall loosening occurs. The differential accumulation of xyloglucan endotransglucosylases, involved in cell wall extensibility (Miedes et al., 2011) and polygalacturonases and pectinases, involved in berry softening (Deytieux-Belleau et al., 2008), are high during berry ripening (Nunan et al., 2001; Lijavetzky et al., 2012). These cell wall modifying genes were remarkably induced in ripe berries with both pre-egressed and egressed B. cinerea, suggesting that the fungus took advantage of the onset of the fruit cell wall self-disassembly, exploiting endogenous developmental programs to activate its own virulence CWDE. In tomato, the expression of the ripening associated genes polygalacturonase and expansin have been shown to facilitate susceptibility to B. cinerea (Cantu et al., 2008; Cantu et al., 2009). It has also been suggested that the fungus can induce unripe fruit cell wall-modifying proteins in order to increase fruit susceptibility (Cantu et al., 2009). We, however, haven’t observed any hastening of ripening process in Botrytis-inoculated samples. Both mock- and Botrytis-inoculated bunches ripen on similar time after inoculation.
Unlike in the flower response to Botrytis infection, in ripe berries with egressed Botrytis we did not observe a regulation of monolignol related genes such as CCoAMT and F5H, involved in cell wall apposition in response to pathogen attack (Busam et al., 1997; Bhuiyan et al., 2009), strongly suggesting there was no cell wall fortification.
In conclusion, B. cinerea inoculated at bloom was quiescent for 12 weeks and egressed at ripening, suggesting that the defense responses of the berries were efficient to halt the fungal growth only until maturity. Our study revealed that the defense responses of immature berries (at 4 wpi) that kept B. cinerea quiescent were similar to the response of the plant to the pathogen at bloom (Haile et al., 2017). During this period, the fungus had cryptic interaction with the berry keeping its basal metabolic activities and deacetylating its cell wall. However, at ripe (at 12 wpi) the pathogen managed to egress and cause bunch rot, using the advantage of the fruit cell wall self-disassembly and fulfillment of other conditions (including humidity). Consequently, there were several defense responses except cell wall strengthening by the ripe berries, but not effective in halting the pathogen from berry colonization.
Data Availability Statement
The datasets generated for this study can be found in the NCBI Short Read Archive, BioProject accession code PRJNA414966.
Author Contributions
ZH conceptualised the project, carried out the lab work and most of the analyses, and drafted the manuscript. GM supported in the interpretation of the data and drafted the manuscript. SP supported in the interpretation of the data. PS, MM, and KE supported in the transcriptomic data analysis. DM and UV carried out the metabolic analysis and supported in the interpretation of the results. EB conceptualised the project and contributed to the discussion of the results. CM conceptualised as well as coordinated the project, contributed to the discussion of the results and to the drafting of the manuscript. All authors read and approved the final manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The research was funded by the Autonomous Province of Trento (PAT-ADP 2013–2016). The authors would like to thank Prof. Paul Tudzynski retired from the Institute for Biology and Biotechnology of Plants (Muenster University) for the availability and the supervision of the work during Haile’s PhD fellowship. Part of the work described in the present manuscript was the object of the Haile’s PhD thesis (Haile, 2017).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2019.01704/full#supplementary-material
References
Abuqamar, S., Chai, M.-F., Luo, H., Song, F., Mengiste, T. (2008). Tomato protein kinase 1b mediates signaling of plant responses to necrotrophic fungi and insect herbivory. Plant Cell 20 (7), 1964–1983. doi: 10.1105/tpc.108.059477
Agudelo-Romero, P., Erban, A., Rego, C., Carbonell-Bejerano, P., Nascimento, T., Sousa, L., et al. (2015). Transcriptome and metabolome reprogramming in Vitis vinifera cv. Trincadeira berries upon infection with Botrytis cinerea. J. Exp. Bot. 66 (7), 1769–1785. doi: 10.1093/jxb/eru517
Alkan, N., Friedlander, G., Ment, D., Prusky, D., Fluhr, R. (2014). Simultaneous transcriptome analysis of Colletotrichum gloeosporioides and tomato fruit pathosystem reveals novel fungal pathogenicity and fruit defense strategies. New Phyt. 205, 801–815. doi: 10.1111/nph.13087
Amselem, J., Cuomo, C. A., van Kan, J. L., Viaud, M., Benito, E. P., Couloux, A., et al. (2011). Genomic analysis of the necrotrophic fungal pathogens Sclerotinia sclerotiorum and Botrytis cinerea. PloS Genet. 7 (8), e1002230. doi: 10.1371/journal.pgen.1002230
Asiimwe, T., Krause, K., Schlunk, I., Kothe, E. (2012). Modulation of ethanol stress tolerance by aldehyde dehydrogenase in the mycorrhizal fungus Tricholoma vaccinum. Mycorrhiza 22 (6), 471–484. doi: 10.1007/s00572-011-0424-9
Barnes, S. E., Shaw, M. W. (2002). Factors affecting symptom production by latent Botrytis cinerea in Primula × polyantha. Plant Path. 51, 746–754. doi: 10.1046/j.1365-3059.2002.00761.x
Bhuiyan, N. H., Selvaraj, G., Wei, Y., King, J. (2009). Gene expression profiling and silencing reveal that monolignol biosynthesis plays a critical role in penetration defence in wheat against powdery mildew invasion. J. Exp. Bot. 60 (2), 509–521. doi: 10.1093/jxb/ern290
Birkenbihl, R. P., Diezel, C., Somssich, I. E. (2012). Arabidopsis WRKY33 is a key transcriptional regulator of hormonal and metabolic responses toward Botrytis cinerea infection. Plant Physiol. 159 (1), 266–285. doi: 10.1104/pp.111.192641
Blanco-Ulate, B., Morales-Cruz, A., Amrine, K. C. H., Labavitch, J. M., Powell, A. L. T., Cantu, D. (2014). Genome-wide transcriptional profiling of Botrytis cinerea genes targeting plant cell walls during infections of different hosts. Front. Plant Sci. 5, 1–16. doi: 10.3389/fpls.2014.00435
Brutus, A., Sicilia, F., Macone, A., Cervone, F., De Lorenzo, G., Lorenzo, G. D. (2010). A domain swap approach reveals a role of the plant wall-associated kinase 1 (WAK1) as a receptor of oligogalacturonides. Proc. Natl. Acad. Sci. U. S. A. 107 (20), 9452–9457. doi: 10.1073/pnas.1000675107
Busam, G., Junghanns, K. T., Kneusel, R. E., Kassemeyer, H. H., Matern, U. (1997). Characterization and expression of caffeoyl-coenzyme A 3-O-methyltransferase proposed for the induced resistance response of Vitis vinifera L. Plant Physiol. 115 (3), 1039–1048. doi: 10.1104/pp.115.3.1039
Cantu, D., Vicente, A. R., Greve, L. C., Dewey, F. M., Bennett, A. B., Labavitch, J. M., et al. (2008). The intersection between cell wall disassembly, ripening, and fruit susceptibility to Botrytis cinerea. Proc. Natl. Acad. Sci. U. S. A. 105 (3), 859–864. doi: 10.1073/pnas.0709813105
Cantu, D., Blanco-Ulate, B., Yang, L., Labavitch, J. M., Bennett, A. B., Powell, A. L. T. (2009). Ripening-regulated susceptibility of tomato fruit to Botrytis cinerea requires NOR but not RIN or ethylene. Plant Physiol. 150 (3), 1434–1449. doi: 10.1104/pp.109.138701
Carisse, O. (2016). “Epidemiology and aerobiology of Botrytis spp,” in Botrytis – the Fungus, the pathogen and its management in agricultural systems. Eds. Fillinger, S., Elad, Y. (Switzerland: Springer International Publishing), 127–148.
Coertze, S., Holz, G. (2002). Epidemiology of Botrytis cinerea on grape: wound infection by dry, airborne conidia. South Afr. J. Enol. Vitic. 23 (2), 72–77. doi: 10.21548/23-2-2157
Commenil, P., Brunet, L., Audran, J. (1997). The development of the grape berry cuticle in relation to susceptibility to bunch rot disease. J. Exp. Bot. 48 (8), 1599–1607. doi: 10.1093/jxb/48.8.1599
Conesa, A., Götz, S., García-Gómez, J. M., Terol, J., Talón, M., Robles, M. (2005). Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21 (18), 3674–3676. doi: 10.1093/bioinformatics/bti610
Cord-Landwehr, S., Melcher, R. L. J., Kolkenbrock, S., Moerschbacher, B. M. (2016). A chitin deacetylase from the endophytic fungus Pestalotiopsis sp. efficiently inactivates the elicitor activity of chitin oligomers in rice cells. Sci. Rep. 6, 38018. doi: 10.1038/srep38018
Dalmais, B., Schumacher, J., Moraga, J., Le Pecheur, P., Tudzynski, B., Collado, I. G., et al. (2011). The Botrytis cinerea phytotoxin botcinic acid requires two polyketide synthases for production and has a redundant role in virulence with botrydial. Mol. Plant Path. 12 (6), 564–579. doi: 10.1111/j.1364-3703.2010.00692.x
De Cremer, K., Mathys, J., Vos, C., Froenicke, L., Michelmore, R. W., Cammue, B. P., et al. (2013). RNAseq-based transcriptome analysis of Lactuca sativa infected by the fungal necrotroph Botrytis cinerea. Plant Cell Environ. 36 (11), 1992–2007. doi: 10.1111/pce.12106
Deising, H., Siegrist, J. (1995). Chitin deacetylase activity of the rust Uromyces viciae-fabae controlled by fungal morphogenesis. FEMS Microbiol. Lett. 127, 207–212. doi: 10.1111/j.1574-6968.1995.tb07475.x
Deytieux-Belleau, C., Vallet, A., Donèche, B., Geny, L. (2008). Pectin methylesterase and polygalacturonase in the developing grape skin. Plant Physiol. Biochem. 46 (7), 638–646. doi: 10.1016/j.plaphy.2008.04.008
Deytieux-Belleau, C., Geny, L., Roudet, J., Mayet, V., Donèche, B., Fermaud, M. (2009). Grape berry skin features related to ontogenic resistance to Botrytis cinerea. Eur. J. Plant Pathol. 125 (4), 551–563. doi: 10.1007/s10658-009-9503-6
Du, Z., Zhou, X., Ling, Y., Zhang, Z., Su, Z. (2010). AgriGO: a GO analysis toolkit for the agricultural community. Nucleic Acids Res. 38, W64–W70. doi: 10.1093/nar/gkq310
Eichorn, K. W., Lorenz, D. H. (1977). Phänologische entwicklungsstadien der rebe. Nachr. Dtsch. Pflanzenschutzd 29, 119–120.
El Gueddari, N. E., Rauchhaus, U., Moerschbacher, B. M., Deising, H. B. (2002). Developmentally regulated conversion of surface-exposed chitin to chitosan in cell walls of plant pathogenic fungi. New Phytol. 156, 103–112. doi: 10.1046/j.1469-8137.2002.00487.x
Elmer, P. A. G., Michailides, T. M. (2004). “Epidemiology of Botrytis cinerea in orchard and vine crops,” in Botrytis: biology, pathology and control. Eds. Elad, Y., Williamson, B., Tudzynski, P., Delan, N. (Dordrecht, the Netherlands: Kluwer Academic), 234–272.
Espino, J. J., Gutiérrez-Sánchez, G., Brito, N., Shah, P., Orlando, R., González, C. (2010). The Botrytis cinerea early secretome. Proteomics 10 (16), 3020–3034. doi: 10.1002/pmic.201000037
Favaron, F., Lucchetta, M., Odorizzi, S., Cunha, A. T. P., Sella, L. (2007). The role of grape polyphenols on trans-resveratrol activity against Botrytis cinerea and of fungal laccase on the solubility of putative grape pr proteins. J. Plant Pathol. 91 (2009), 579–588. doi: 10.4454/jpp.v91i3.549
Frías, M., González, C., Brito, N. (2011). BcSpl1, a cerato-platanin family protein, contributes to Botrytis cinerea virulence and elicits the hypersensitive response in the host. New Phytol. 192 (2), 483–495. doi: 10.1111/j.1469-8137.2011.03802.x
Frías, M., Brito, N., González, M., González, C. (2014). The phytotoxic activity of the cerato-platanin BcSpl1 resides in a two-peptide motif on the protein surface. Mol. Plant Pathol. 15 (4), 342–351. doi: 10.1111/mpp.12097
Gao, Q.-M., Venugopal, S., Navarre, D., Kachroo, A. (2011). Low oleic acid-derived repression of jasmonic acid-inducible defense responses requires the WRKY50 and WRKY51 proteins. Plant Physiol. 155 (1), 464–476. doi: 10.1104/pp.110.166876
Garrett-Engele, P., Moilanen, B., Cyert, M. S. (1995). Calcineurin, the Ca2+/calmodulin- dependent protein phosphatase, is essential in yeast mutants with cell integrity defects and in mutants that lack a functional vacuolar H(+)-ATPase. Mol. Cell. Biol. 15 (8), 4103–4114. doi: 10.1128/mcb.15.8.4103
Goetz, G., Fkyerat, A., Metais, N., Kunz, M., Tabacchi, R., Pezet, R., et al. (1999). Resistance factors to grey mould in grape berries: identification of some phenolics inhibitors of Botrytis cinerea stilbene oxidase. Phytochemistry 52 (5), 759–767. doi: 10.1016/S0031-9422(99)00351-9
Grimplet, J., Van Hemert, J., Carbonell-Bejerano, P., Díaz-Riquelme, J., Dickerson, J., Fennell, A., et al. (2012). Comparative analysis of grapevine whole-genome gene predictions, functional annotation, categorization and integration of the predicted gene sequences. BMC Res. Notes 5, 213. doi: 10.1186/1756-0500-5-213
Guidarelli, M., Zoli, L., Orlandini, A., Bertolini, P., Baraldi, E. (2014). The mannose-binding lectin gene FaMBL1 is involved in the resistance of unripe strawberry fruits to Colletotrichum acutatum. Mol. Plant Pathol. 15, 832–840. doi: 10.1111/mpp.12143
Gutterson, N., Reuber, T. L. (2004). Regulation of disease resistance pathways by AP2/ERF transcription factors. Curr. Opin. Plant Biol. 7 (4), 465–471. doi: 10.1016/j.pbi.2004.04.007
Höll, J., Vannozzi, A., Czemmel, S., D'Onofrio, C., Walker, A. R., Rausch, T., et al. (2013). The R2R3-MYB transcription factors MYB14 and MYB15 regulate stilbene biosynthesis in Vitis vinifera. Plant Cell 25 (10), 4135–4149. doi: 10.1105/tpc.113.117127
Haile, Z. M., Malacarne, G., Pilati, S., Sonego, P., Engelen, K., Lionetti, V., et al. (2019). The molecular dialogue between grapevine inflorescence/berry and Botrytis cinerea during initial, quiescent and egression infection stages. Acta Hortic. 1248, 587–594. doi: 10.17660/ActaHortic.2019.1248.79
Haile, Z. M. (2017). A molecular study of the Botrytis cinerea–Vitis vinifera interaction: from inflorescence to ripe berry. [PhD’s thesis] (Bologna: University of Bologna).
Haile, Z. M., Pilati, S., Sonego, P., Malacarne, G., Vrhovsek, U., Engelen, K., et al. (2017). Molecular analysis of the early interaction between the grapevine flower and Botrytis cinerea reveals that prompt activation of specific host pathways leads to fungus quiescence. Plant Cell Environ. 40 (8), 1409–1428. doi: 10.1111/pce.12937
Hammerbacher, A., Ralph, S. G., Bohlmann, J., Fenning, T. M., Gershenzon, J., Schmidt, A. (2011). Biosynthesis of the major tetrahydroxystilbenes in spruce, astringin and isorhapontin, proceeds via resveratrol and is enhanced by fungal infection. Plant Physiol. 157 (2), 876–890. doi: 10.1104/pp.111.181420
Hellemans, J., Mortier, G., De Paepe, A., Speleman, F., Vandesompele, J. (2007). qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol. 8 (2), R19. doi: 10.1186/gb-2007-8-2-r19
Jeandet, P., Bessis, R., Sbaghi, M., Meunier, P. (1995). Production of the phytoalexin resveratrol by grapes as a response to Botrytis attack under natural conditions. J. Phytopathol. 143, 135–139. doi: 10.1111/j.1439-0434.1995.tb00246.x
Jersch, S., Scherer, C., Huth, G., Schlösser, E. (1989). Proanthocyanidins as basis for quiescence of Botrytis cinerea in immature strawberry fruits. Z. Pflanzenkr. Pflanzenschutz 96, 365–378.
Jiang, Y., Deyholos, M. K. (2009). Functional characterization of Arabidopsis NaCl- inducible WRKY25 and WRKY33 transcription factors in abiotic stresses. Plant Mol. Biol. 69 (1–2), 91–105. doi: 10.1007/s11103-008-9408-3
Keller, M., Viret, O., Cole, F. M. (2003). Botrytis cinerea infection in grape flowers: defense reaction, latency, and disease expression. Phytopathology 93 (2), 316–322. doi: 10.1094/Phyto.2003.93.3.316
Kelloniemi, J., Trouvelot, S., Héloir, M. C., Simon, A., Dalmais, B., Frettinger, P., et al. (2015). Analysis of the molecular dialogue between gray mold (Botrytis cinerea) and grapevine (Vitis vinifera) reveals a clear shift in defense mechanisms during berry ripening. Mol. Plant Microbe Interact. 28 (11), 1167–1180. doi: 10.1094/MPMI-02-15-0039-R
Kretschmer, M., Kassemeyer, H.-H., Hahn, M. (2007). Age-dependent grey mould susceptibility and tissue-specific defence gene activation of grapevine berry skins after infection by Botrytis cinerea. J. Phytopathol. 155 (5), 258–263. doi: 10.1111/j.1439-0434.2007.01216.x
Kwon, S. J., Jin, H. C., Lee, S., Nam, M. H., Chung, J. H., Kwon, S., et al. (2009). GDSL lipase-like 1 regulates systemic resistance associated with ethylene signaling in Arabidopsis. Plant J. 58 (2), 235–245. doi: 10.1111/j.1365-313X.2008.03772.x
Langcake, P. (1981). Disease resistance of Vitis spp. and the production of the stress metabolites resveratrol, ϵ-viniferin, α-viniferin and pterostilbene. Physiol. Plant Pathol. 18 (2), 213–226. doi: 10.1016/S0048-4059(81)80043-4
Law, C. W., Chen, Y., Shi, W., Smyth, G. K. (2014). voom: precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 15 (2), R29. doi: 10.1186/gb-2014-15-2-r29
Li, S., Fu, Q., Chen, L., Huang, W., Yu, D. (2011). Arabidopsis thaliana WRKY25, WRKY26, and WRKY33 coordinate induction of plant thermotolerance. Planta 233 (6), 1237–1252. doi: 10.1007/s00425-011-1375-2
Liao, Y., Smyth, G. K., Shi, W. (2013). The Subread aligner: fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res. 41 (10), e108. doi: 10.1093/nar/gkt214
Liao, Y., Smyth, G. K., Shi, W. (2014). FeatureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30 (7), 923–930. doi: 10.1093/bioinformatics/btt656
Licausi, F., Ohme-Takagi, M., Perata, P. (2013). APETALA2/ethylene responsive factor (AP2/ERF) transcription factors: mediators of stress responses and developmental programs. New Phytol. 199 (3), 639–649. doi: 10.1111/nph.12291
Lijavetzky, D., Carbonell-Bejerano, P., Grimplet, J., Bravo, G., Flores, P., Fenoll, J., et al. (2012). Berry flesh and skin ripening features in Vitis vinifera as assessed by transcriptional profiling. PloS One 7 (6), e39547. doi: 10.1371/journal.pone.0039547
Lionetti, V., Cervone, F., Bellincampi, D. (2012). Methyl esterification of pectin plays a role during plant–pathogen interactions and affects plant resistance to diseases. J. Plant Physiol. 169, 1623–1630. doi: 10.1016/j.jplph.2012.05.006
Liu, T., Liu, Z., Song, C., Hu, Y., Han, Z., She, J., et al. (2012). Chitin-induced dimerization activates a plant immune receptor. Science 336 (6085), 1160–1164. doi: 10.1126/science.1218867
Malacarne, G., Vrhovsek, U., Zulini, L., Cestaro, A., Stefanini, M., Mattivi, F., et al. (2011). Resistance to Plasmopara viticola in a grapevine segregating population is associated with stilbenoid accumulation and with specific host transcriptional responses. BMC Plant Biol. 11 (1), 114. doi: 10.1186/1471-2229-11-114
Martin, M. (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17, 10–12. doi: 10.14806/ej.17.1.200
McClellan, W. D., Hewitt, W. B. (1973). Early Botrytis rot of grape time of infection and latency of Bc in Vitis vinifera. Phytopathology 63, 1151–1157. doi: 10.1094/Phyto-63-1151
McNicol, R. J., Williamson, B. (1989). Systemic infection of black currant flowers by Botrytis cinerea and its possible involvement in premature abscission of fruits. Ann. Appl. Biol. 114 (2), 243–254. doi: 10.1111/j.1744-7348.1989.tb02101.x
Merz, P. R., Moser, T., Höll, J., Kortekamp, A., Buchholz, G., Zyprian, E., et al. (2015). The transcription factor VvWRKY33 is involved in the regulation of grapevine (Vitis vinifera) defense against the oomycete pathogen Plasmopara viticola. Physiol. Plant, 153 (3), 365–380. doi: 10.1111/ppl.12251
Micheli, F. (2001). Pectin methylesterases: cell wall enzymes with important roles in plant physiology. Trends Plant Sci. 6 (9), 414–419. doi: 10.1016/S1360-1385(01)02045-3
Miedes, E., Zarra, I., Hoson, T., Herbers, K., Sonnewald, U., Lorences, E. P. (2011). Xyloglucan endotransglucosylase and cell wall extensibility. J. Plant Physiol. 168 (3), 196–203. doi: 10.1016/j.jplph.2010.06.029
Mlikota-Gabler, F., Smilanick, J. L., Mansour, M., Ramming, D. W., Mackey, B. E. (2003). Correlations of morphological, anatomical, and chemical features of grape berries with resistance to Botrytis cinerea. Phytopathology 93 (10), 1263–1273. doi: 10.1094/PHYTO.2003.93.10.1263
Monteiro, S., Barakat, M., Piçarra-Pereira, M. A., Teixeira, A. R., Ferreira, R. B. (2003). Osmotin and thaumatin from grape: a putative general defense mechanism against pathogenic fungi. Phytopathology 93 (12), 1505–1512. doi: 10.1094/PHYTO.2003.93.12.1505
Moretto, M., Sonego, P., Dierckxsens, N., Brilli, M., Bianco, L., Ledezma-Tejeida, D., et al. (2016). COLOMBOS v3.0: leveraging gene expression compendia for cross-species analyses. Nucleic Acids Res. 44 (D1), D620–D623. doi: 10.1093/nar/gkv1251
Nair, N., Guilbaud-Oulton, S., Barchia, I., Emmet, R., G, N., Nair, N., et al. (1995). Significance of carry over inoculum, flower infection and latency on the incidence of Botrytis cinerea in berries of grapevines at harvest in New South Wales. Aust. J. Exp. Agr. 35, 1177–1180. doi: 10.1071/EA9951177
Nakajima, M., Akutsu, K. (2014). Virulence factors of Botrytis cinerea. J. Gen. Plant Pathol. 80 (1), 15–23. doi: 10.1007/s10327-013-0492-0
Nunan, K. J., Davies, C., Robinson, S. P., Fincher, G. B. (2001). Expression patterns of cell wall-modifying enzymes during grape berry development. Planta 214 (2), 257–264. doi: 10.1007/s004250100609
Oh, I. S., Park, A. R., Bae, M. S., Kwon, S. J., Kim, Y. S., Lee, J. E., et al. (2005). Secretome analysis reveals an Arabidopsis lipase involved in defense against Alternaria brassicicola. Plant Cell 17 (10), 2832–2847. doi: 10.1105/tpc.105.034819
Oirdi, M. El, Abd, T., Rahman, E., Rigano, L., Hadrami, A. El, Daayf, F., et al. (2011). Botrytis cinerea manipulates the antagonistic effects between immune pathways to promote disease development in tomato. Plant Cell 23, 2405–2421. doi: 10.1105/tpc.111.083394
Oliveros, J. C. (2015). Venny. An interactive tool for comparing lists with Venn’s diagrams. 2007-2015. https://bioinfogp.cnb.csic.es/tools/venny/index.html.
Padgett, M., Morrison, J. C. (1990). Changes in grape berry exudates during fruit development and their effect on mycelial growth of Botrytis cinerea. J. Am. Soc Hortic. Sci. 115 (2), 269–273. doi: 10.21273/JASHS.115.2.269
Paré, A., Kim, M., Juarez, M. T., Brody, S., McGinnis, W. (2012). The functions of grainy head-like proteins in animals and fungi and the evolution of apical extracellular barriers. PloS One 7 (5), e36254. doi: 10.1371/journal.pone.0036254
Pardo, M., Monteoliva, L., Vázquez, P., Martínez, R., Molero, G., Nombela, C., et al. (2004). PST1 and ECM33 encode two yeast cell surface GPI proteins important for cell wall integrity. Microbiology 150 (12), 4157–4170. doi: 10.1099/mic.0.26924-0
Pernas, M., López-Solanilla, E., Sánchez-Monge, R., Salcedo, G., Rodríguez-Palenzuela, P. (1999). Antifungal activity of a plant cystatin. Mol. Plant Microbe Interact. 12 (7), 624–627. doi: 10.1094/MPMI.1999.12.7.624
Petutschnig, E. K., Jones, A. M. E., Serazetdinova, L., Lipka, U., Lipka, V. (2010). The lysin motif receptor-like kinase (LysM-RLK) CERK1 is a major chitin-binding protein in Arabidopsis thaliana and subject to chitin-induced phosphorylation. J. Biolog. Chem. 285 (37), 28902–28911. doi: 10.1074/jbc.M110.116657
Pezet, R., Perret, C., Jean-Denis, J. B., Tabacchi, R., Gindro, K., Viret, O. (2003a). δ-viniferin, a resveratrol dehydrodimer: one of the major stilbenes synthesized by stressed grapevine leaves. J. Agr. Food Chem. 51 (18), 5488–5492. doi: 10.1021/jf030227o
Pezet, R., Viret, O., Perret, C., Tabacchi, R. (2003b). Latency of Botrytis cinerea Pers.: Fr. and biochemical studies during growth and ripening of two grape berry cultivars, respectively susceptible and resistant to grey mould. Phytopathol 214, 208–214. doi: 10.1046/j.1439-0434.2003.00707.x
Prusky, D., Alkan, N., Mengiste, T., Fluhr, R. (2013). Quiescent and necrotrophic lifestyle choice during postharvest disease development. Ann. Rev. Phytopathol. 51, 155–176. doi: 10.1146/annurev-phyto-082712-102349
Prusky, D. (1996). Pathogen quiescence in postharvest diseases. Ann. Rev. Phytopathol. 34, 413–434. doi: 10.1146/annurev.phyto.34.1.413
Rieu, I., Powers, S. J. (2009). Real-time quantitative RT-PCR: design, calculations, and statistics. Plant Cell 21 (4), 1031–1033. doi: 10.1105/tpc.109.066001
Rolke, Y., Liu, S., Quidde, T., Williamson, B., Schouten, A., Weltring, K.-M., et al. (2004). Functional analysis of H2O2-generating systems in Botrytis cinerea: the major Cu–Zn–superoxide dismutase (BCSOD1) contributes to virulence on French bean, whereas a glucose oxidase (BCGOD1) is dispensible. Mol. Plant Pathol. 5 (1), 17–27. doi: 10.1046/J.1364-3703.2004.00201.X
Ruijter, J. M., Ramakers, C., Hoogaars, W. M. H., Karlen, Y., Bakker, O., van den hoff, M. J. B., et al. (2009). Amplification efficiency: linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Res. 37 (6), e45. doi: 10.1093/nar/gkp045
Saga, H., Ogawa, T., Kai, K., Suzuki, H., Ogata, Y., Sakurai, N., et al. (2012). Identification and characterization of ANAC042, a transcription factor family gene involved in the regulation of camalexin biosynthesis in Arabidopsis. Mol. Plant Microbe Interact. 25 (5), 684–696. doi: 10.1094/MPMI-09-11-0244
Schouten, A., Wagemakers, L., Stefanato, F. L., van der Kaaij, R. M., van Kan, J. A. L. (2002). Resveratrol acts as a natural profungicide and induces self-intoxication by a specific laccase. Mol. Microbiol. 43 (4), 883–894. doi: 10.1046/j.1365-2958.2002.02801.x
Schumacher, J., Simon, A., Cohrs, K. C., Traeger, S., Porquier, A., Dalmais, B., et al. (2015). The VELVET complex in the gray mold fungus Botrytis cinerea: impact of BcLAE1 on differentiation, secondary metabolism, and virulence. Mol. Plant Microbe Interact. 28 (6), 659–674. doi: 10.1094/MPMI-12-14-0411-R
Shaw, M. W., Emmanuel, C. J., Emilda, D., Terhem, R. B., Shafia, A., Tsamaidi, D., et al. (2016). Analysis of cryptic, systemic Botrytis infections in symptomless hosts. Front. Plant Sci. 7, 625. doi: 10.3389/fpls.2016.00625
Siewers, V., Viaud, M., Jimenez-Teja, D., Collado, I. G., Gronover, C. S., Pradier, J.-M., et al. (2005). Functional analysis of the cytochrome p450 monooxygenase gene bcbot1 of Botrytis cinerea indicates that botrydial is a strain-specific virulence factor. Mol. Plant Microbe Interact. 602 (6), 602–612. doi: 10.1094/MPMI-18-0602
Smith, J. E., Mengesha, B., Tang, H., Mengiste, T., Bluhm, B. H. (2014). Resistance to Botrytis cinerea in Solanum lycopersicoides involves widespread transcriptional reprogramming. BMC Genom. 15, 334. doi: 10.1186/1471-2164-15-334
Smyth, G. K. (2004). Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 3 (1), 1–25. doi: 10.2202/1544-6115.1027
Supek, F., Bošnjak, M., Škunca, N., Šmuc, T. (2011). Revigo summarizes and visualizes long lists of gene ontology terms. PloS One 6 (7), e21800. doi: 10.1371/journal.pone.0021800
ten Have, A., Espino, J. J., Dekkers, E., Van Sluyter, S. C., Brito, N., Kay, J., et al. (2010). The Botrytis cinerea aspartic proteinase family. Fungal Genet. Biol. 47 (1), 53–65. doi: 10.1016/j.fgb.2009.10.008
Thimm, O., Bläsing, O., Gibon, Y., Nagel, A., Meyer, S., Krüger, P., et al. (2004). MAPMAN: A user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 37 (6), 914–939. doi: 10.1111/j.1365313X.2004.02016.x
van Kan, J. A. L., Stassen, J. H. M., Mosbach, A., Van Der Lee, T. A. J., Faino, L., Farmer, A. D., et al. (2016). A gapless genome sequence of the fungus Botrytis cinerea. Mol. Plant Pathol. 8, 75–89. doi: 10.1111/mpp.12384
Vandesompele, J., De Preter, K., Pattyn, F., Poppe, B., Van Roy, N., De Paepe, A., et al. (2002). Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3 (7), RESEARCH0034. doi: 10.1186/gb-2002-3-7-research0034
Wang, Q., Song, C., Li, C. C. H. (2004). Molecular perspectives on p97-VCP: progress in understanding its structure and diverse biological functions. J. Struct. Biol. 146 (1–2), 44–57. doi: 10.1016/j.jsb.2003.11.014
Wang, D., Amornsiripanitch, N., Dong, X. (2006). A genomic approach to identify regulatory nodes in the transcriptional network of systemic acquired resistance in plants. PloS Pathog. 2 (11), 1042–1050. doi: 10.1371/journal.ppat.0020123
Williamson, B. Y. B., Mcnicol, R. J., Dolan, A., Crop, S., Do, D. (1987). The effect of inoculating flowers and developing fruits with Botrytis cinerea on post-harvest grey mould of red raspberry. Ann. Appl. Biol. 111, 285–294. doi: 10.1111/j.1744-7348.1987.tb01454.x
Keywords: Botrytis cinerea, hard-green berry, ripening, egression, quiescence, Vitis vinifera
Citation: Haile ZM, Malacarne G, Pilati S, Sonego P, Moretto M, Masuero D, Vrhovsek U, Engelen K, Baraldi E and Moser C (2020) Dual Transcriptome and Metabolic Analysis of Vitis vinifera cv. Pinot Noir Berry and Botrytis cinerea During Quiescence and Egressed Infection. Front. Plant Sci. 10:1704. doi: 10.3389/fpls.2019.01704
Received: 04 October 2019; Accepted: 04 December 2019;
Published: 30 January 2020.
Edited by:
Brigitte Mauch-Mani, Université de Neuchâtel, SwitzerlandReviewed by:
Vittorio Rossi, Catholic University of the Sacred Heart, ItalySilvia Proietti, Università degli Studi della Tuscia, Italy
Copyright © 2020 Haile, Malacarne, Pilati, Sonego, Moretto, Masuero, Vrhovsek, Engelen, Baraldi and Moser. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Giulia Malacarne, giulia.malacarne@fmach.it
 Zeraye Mehari Haile
Zeraye Mehari Haile Giulia Malacarne
Giulia Malacarne Stefania Pilati
Stefania Pilati Paolo Sonego
Paolo Sonego Marco Moretto
Marco Moretto Domenico Masuero
Domenico Masuero Urska Vrhovsek
Urska Vrhovsek Kristof Engelen6
Kristof Engelen6 Elena Baraldi
Elena Baraldi Claudio Moser
Claudio Moser