- 1Department of Marine Science, Marine Science Institute, University of Texas at Austin, Port Aransas, TX, United States
- 2Earth, Marine and Environmental Sciences, University of North Carolina at Chapel Hill, Chapel Hill, NC, United States
- 3Organic Geochemistry Group, MARUM-Center for Marine Environmental Sciences and Department of Geosciences, University of Bremen, Bremen, Germany
The production and anaerobic oxidation of methane (AOM) by microorganisms is widespread in organic-rich deep subseafloor sediments. Yet, the organisms that carry out these processes remain largely unknown. Here we identify members of the methane-cycling microbial community in deep subsurface, hydrate-containing sediments of the Peru Trench by targeting functional genes of the alpha subunit of methyl coenzyme M reductase (mcrA). The mcrA profile reveals a distinct community zonation that partially matches the zonation of methane oxidizing and –producing activity inferred from sulfate and methane concentrations and carbon-isotopic compositions of methane and dissolved inorganic carbon (DIC). McrA appears absent from sulfate-rich sediments that are devoid of methane, but mcrA sequences belonging to putatively methane-oxidizing ANME-1a-b occur from the zone of methane oxidation to several meters into the methanogenesis zone. A sister group of ANME-1a-b, referred to as ANME-1d, and members of putatively aceticlastic Methanothrix (formerly Methanosaeta) occur throughout the remaining methanogenesis zone. Analyses of 16S rRNA and mcrA-mRNA indicate that the methane-cycling community is alive throughout (rRNA to 230 mbsf) and active in at least parts of the sediment column (mRNA at 44 mbsf). Carbon-isotopic depletions of methane relative to DIC (−80 to −86‰) suggest mostly methane production by CO2 reduction and thus seem at odds with the widespread detection of ANME-1 and Methanothrix. We explain this apparent contradiction based on recent insights into the metabolisms of both ANME-1 and Methanothricaceae, which indicate the potential for methanogenetic growth by CO2 reduction in both groups.
Introduction
The detection of active microbial populations to 80 mbsf in Peru Margin sediments during Ocean Drilling Program (ODP) Leg 112 in 1988, was the first demonstration of an deep subseafloor biosphere (Cragg et al., 1990). Since then, numerous studies and multiple lines of evidence from a range of locations have shown a vast microbial biomass in deep subseafloor sediments (for syntheses, see D’Hondt et al., 2004; Kallmeyer et al., 2012; Parkes et al., 2014) with metabolically active cells to at least 1,500 mbsf (Roussel et al., 2008; Inagaki et al., 2015; Heuer et al., 2020), and the existence of a subsurface microbiome that is distinct from that found in marine surface sediments (e.g., Deng et al., 2020; Hoshino et al., 2020).
Several sites sampled during ODP Leg 112 were revisited in 2002 during ODP Leg 201, now 22 years ago, during the first ocean drilling expedition to focus on subseafloor life (D’Hondt et al., 2003). Porewater concentration gradients of microbially consumed electron acceptors such as nitrate or sulfate indicated active microbial populations to depths of >400 mbsf in the sediment column (D’Hondt et al., 2004). Molecular biological studies, e.g., polymerase-chain-reaction (PCR) assays of 16S rRNA genes (Parkes et al., 2005; Inagaki et al., 2006; Webster et al., 2006) and 16S rRNA gene transcripts (Biddle et al., 2006; Sørensen and Teske, 2006), fluorescence-in-situ-hybridization (FISH; Mauclaire et al., 2005, Schippers et al., 2005), and metagenomic signatures of whole-genome amplified DNA (Biddle et al., 2008) provided insights into the community structure and metabolic potential of microbial populations. Yet, specific links between microbial activity based on geochemical gradients and microbial identity based on genetic and genomic assays could not be established. For instance, sulfate and methane profiles suggested that sulfate reduction, anaerobic oxidation of methane (AOM), and methanogenesis were all important microbially-driven in situ processes (D’Hondt et al., 2004). However, sulfate-reducing, methanogenic, or methane-oxidizing microorganisms were surprisingly rare or absent from clone libraries of transcribed, PCR-amplified 16S rRNA (Biddle et al., 2006; Sørensen and Teske, 2006) and PCR-amplified 16S rRNA genes (Parkes et al., 2005; Inagaki et al., 2006).
Functional genes that encode for enzymes that are unique to certain metabolisms can be targeted to identify microorganisms that are involved in these metabolisms. Functional genes that have been investigated in targeted studies at ODP Leg 201 sites include the gene for dissimilatory sulfite reductase (dsrAB), a key enzyme of dissimilatory sulfate reduction (Wagner et al., 2005), the gene for reductive dehalogenase (rdhA) of reductive dehalorespiration (Futagami et al., 2009), the gene for formyl tetrahydrofolate synthetase (fhsA), a crucial enzyme of acetogenewsis (Lever et al., 2010), and the gene for the α subunit of methyl coenzyme M reductase (mcrA), an enzyme that catalyzes the terminal step of biological methanogenesis and is also present in anaerobic methane oxidizers (Friedrich, 2005; Knittel and Boetius, 2009; Wang et al., 2021). Patchy PCR detections of dsrAB and mcrA in only a few samples (Parkes et al., 2005; Inagaki et al., 2006; Webster et al., 2006) remain at odds with porewater concentration profiles of sulfate and methane, which indicate microbial sulfate reduction, AOM, and methanogenesis (D’Hondt et al., 2004). Similar observations were made based on quantitative PCR and metagenome sequencing in methane-rich deep subseafloor sediments of Hydrate Ridge in the Northeastern Pacific (Colwell et al., 2008), the Black Sea and off Namibia (Schippers et al., 2012), the Baltic Sea (Marshall et al., 2018), and Adélie Basin off Antarctica (Carr et al., 2018). It was thus proposed that methanogens account for low percentages (<1%) of microbial cells in subseafloor sediments, or are not detected by PCR assays due to primer mismatches or use of unrecognized genetic pathways (Lever, 2013).
Here we take a closer look at the in situ community of methanogens and anaerobic methanotrophs in the sediment column of ODP Site 1230 in the Peru Trench via PCR assays of mcrA. We investigate the relationship between community zonation and geochemical profiles [sulfate, methane, formate, acetate, hydrogen, δ13C-methane and -dissolved inorganic carbon (DIC)], and identify active members of the methane-cycling community via reverse transcription-PCR (RT-PCR) of 16S rRNA and mcrA-mRNA. Redesigned general mcrA primers (Lever and Teske, 2015) and new group-specific mcrA and 16S rRNA gene primers allow us to detect methane-cycling functional genes in the AOM and methanogenesis zones inferred from porewater chemical gradients. While updated primers improve the detection of methane-cycling archaea, they reinforce the notion that methane-cycling archaea only account for a small proportion of microbial subsurface communities even in sediments with clear geochemical evidence for methanogenesis and AOM.
Materials and methods
Field site and sampling
The Peru Trench is part of the larger Atacama Trench that is located between the continental South American Plate and the accretionary wedge of the oceanic Nazca Plate (Suess, 1981). ODP Site 1230 is located on the lower slope of the Peru Trench at 5,086 m water depth (Figure 1). Sediments were drilled to ~270 mbsf during ODP Leg 201 in 2002 (D’Hondt et al., 2003). Three boreholes (A, B, and C) were within ~20 m of one another (D’Hondt et al., 2003). Sediment temperatures are low, increasing linearly from 2°C at the seafloor to 12°C at 270 mbsf. The upper 200 m of sediment consist of clay-rich, diatomaceous mud that was largely relocated from the continental shelf throughout the Holocene and Pleistocene. At approximately 216 mbsf, the sediment column changes to Miocene diatom ooze in a stratigraphic hiatus of 4.5 million years (Shipboard Scientific Party, 1988; Meister et al., 2005). Throughout the sediment column, organic carbon contents mostly range from 2 to 4% dry sediment weight (Meister et al., 2005). DIC concentrations to 160 mM and sulfate depletion in the upper ~10 mbsf indicate active microbial remineralization of organic matter, largely by sulfate reduction (D’Hondt et al., 2003). After sulfate is depleted, methane concentrations increase rapidly and reach in situ saturation by 28 mbsf (Spivack et al., 2005). Geophysical and chemical data suggest that hydrates are first present at ~70 mbsf and occur intermittently to 278 mbsf (D’Hondt et al., 2003).
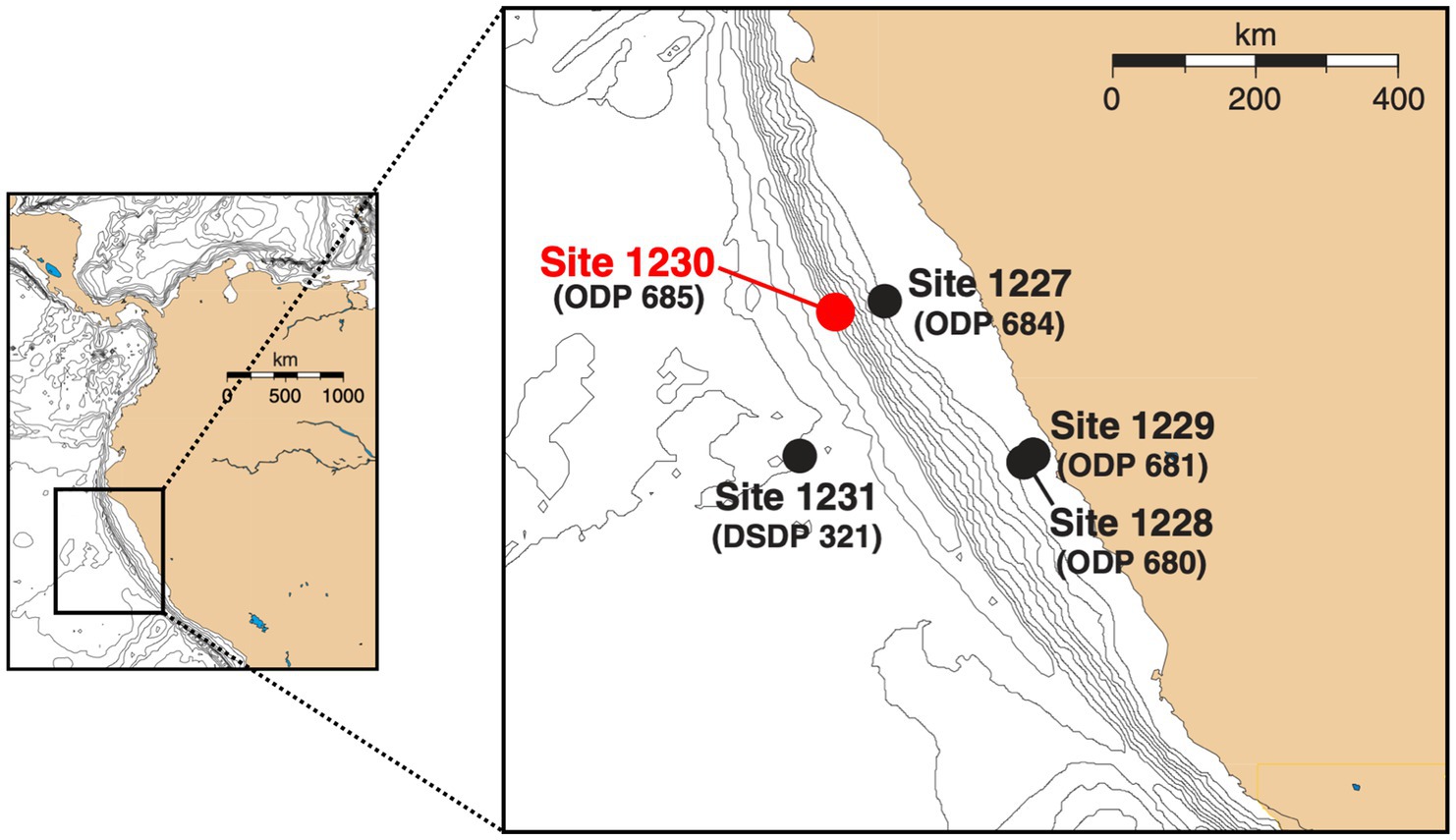
Figure 1. Map of Peru Margin sites sampled during ODP Leg 201 and ODP Leg 112 (in parentheses) [adapted from D’Hondt et al., 2003]. Samples used in this study were collected at ODP Site 1230 in the Peru Trench, which was in the same location as the previously studied ODP Site 685.
For molecular biological analyses, 5-cm whole-round intervals of cores were frozen at −80°C. Only sediment from the nearly contamination-free core interiors was used (House et al., 2003; Lever et al., 2006). For carbon isotope analyses, 5-mL subsamples were frozen in pre-combusted glass vials.
Porewater geochemical concentrations
We used published depth profiles of DIC, sulfate, methane, dihydrogen (H2), formate and acetate concentrations (Figures 2A–E; D’Hondt et al., 2003). Due to outgassing during core retrieval, measured methane concentrations below ~12 mbsf were underestimates of in situ concentrations. We calculated methane concentrations below the saturation depth at in situ temperature, pressure, and salinity, assuming a uniform pore size of 1.0 μm based on the equilibrium model for methane hydrate-seawater-porous media (Sun and Duan, 2007). Modeled methane concentrations generally agree with measured in situ methane concentrations based on pressure coring (4 depths analyzed at ODP Site 1230; Spivack et al., 2005).
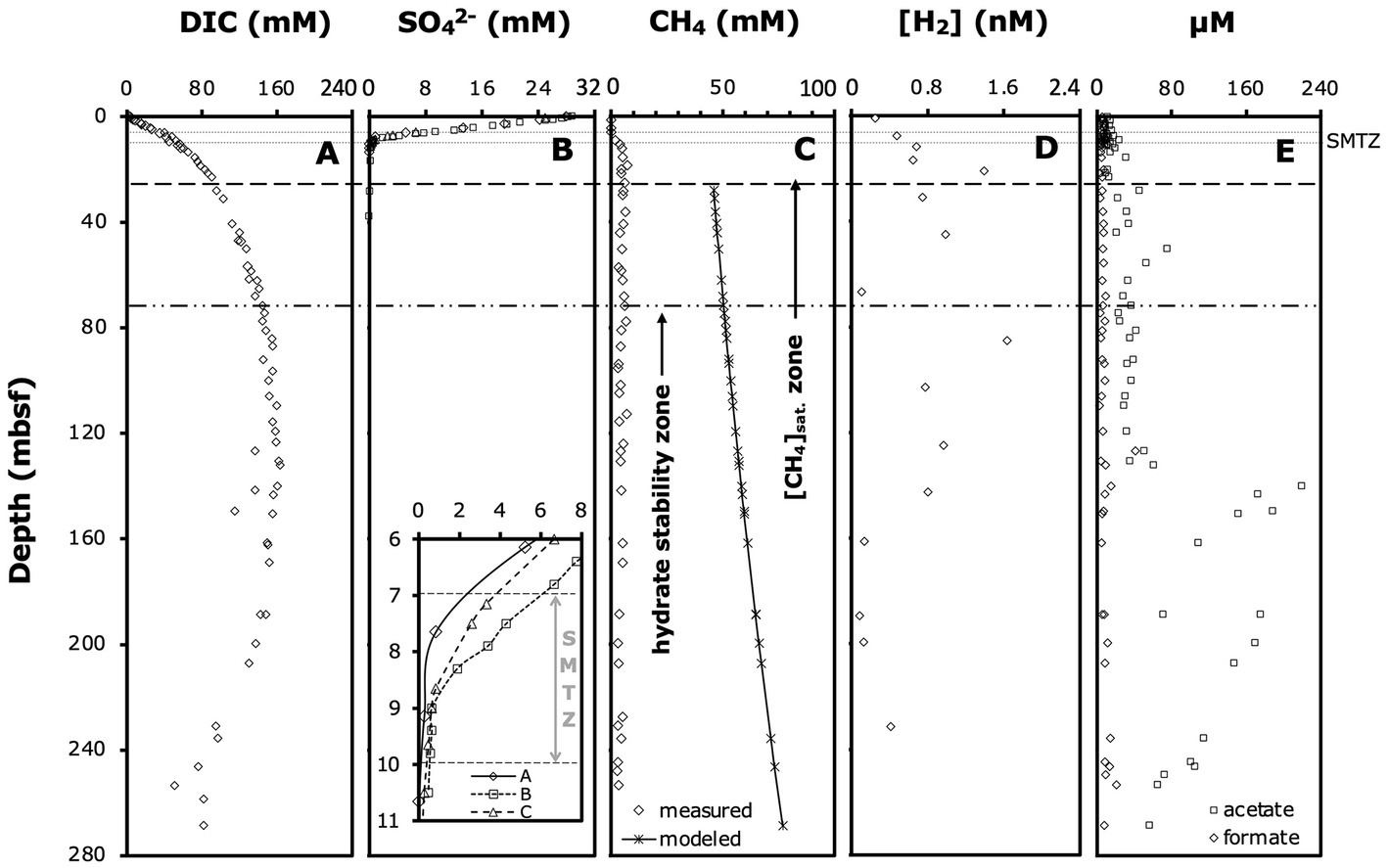
Figure 2. Relevant porewater geochemical profiles: (A) DIC concentrations, (B) sulfate concentrations (note: the insert shows an enlarged view of the SMTZ and concentrations in individual boreholes), (C) measured methane concentrations, modeled methane saturation concentrations, and distribution of hydrate stability zone (D’Hondt et al., 2003) and methane saturation zone (Spivack et al., 2005), (D) dihydrogen (H2) concentrations, and (E) formate and acetate concentrations. Modeled methane concentrations from this study, all other data from D’Hondt et al. (2003). Horizontal lines indicate the approximate depth interval of the SMTZ, and the depths below which we estimate methane concentrations to be saturated and methane hydrates to be present. All data are from Borehole A, except where noted.
δ13C-C1 and DIC
δ13C-C1 (~99% 13C-CH4) and -DIC (Figure 3) were measured as described previously (Biddle et al., 2006). All values are shown in Supplementary Table S1.
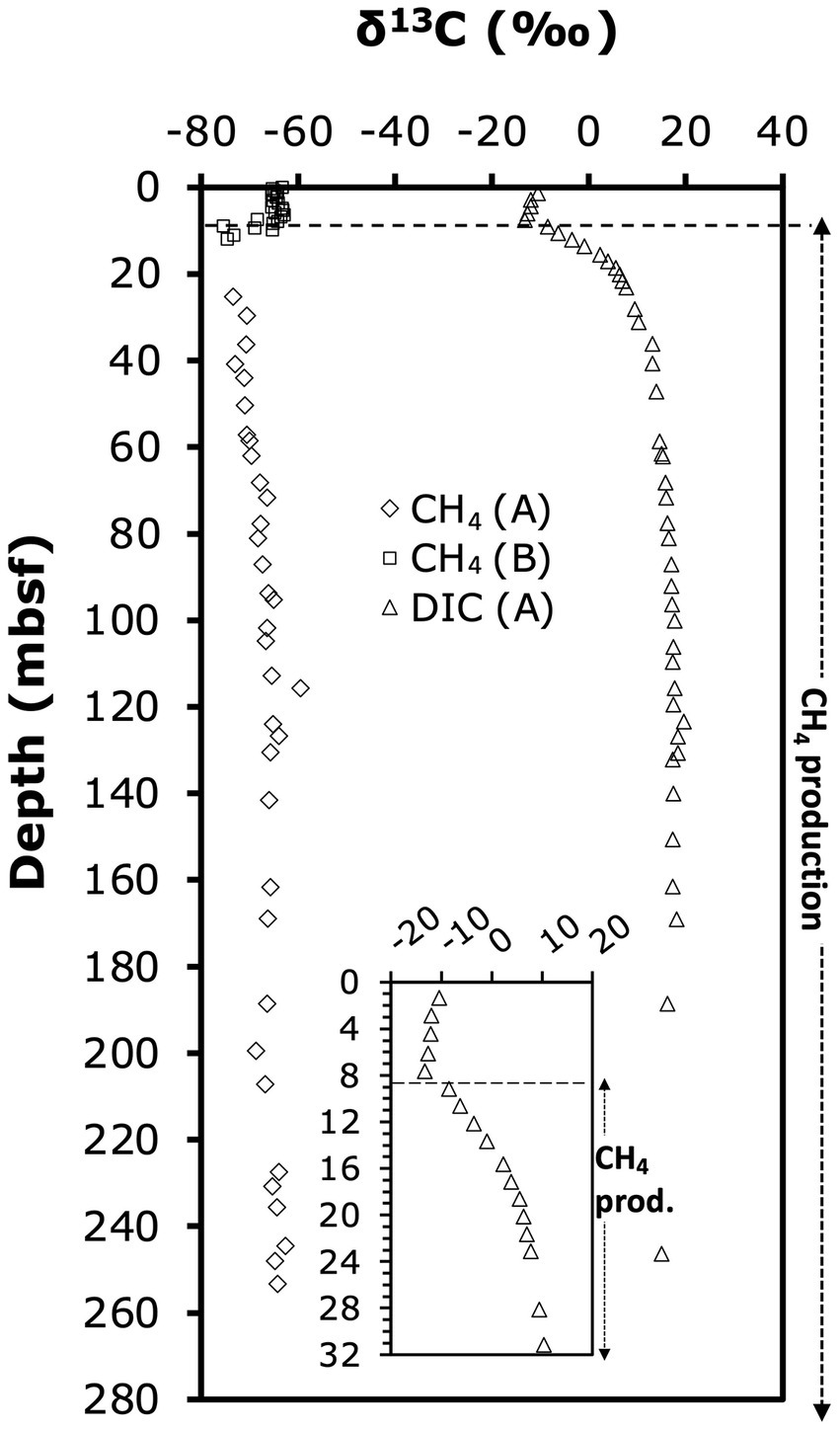
Figure 3. Depth profile of δ13C-CH4 and δ13C-DIC. Letters in parentheses indicate borehole in which samples were analyzed. Depth profile of difference between δ13C-CH4 and δ13C-DIC. The dashed line marks the abrupt shift in δ13C-DIC from decreasing to increasing values.
Nucleic acid extraction
RNA was extracted as in Biddle et al. (2006), except that the extraction buffer was supplemented with 120 mM sodium phosphate. DNA was extracted using the same protocol as for RNA, except that the pH of the extraction buffer and phenol were raised to 8.0, the bead beating time reduced to 15 s, and the bead beating speed reduced to 4.0 (Qbiogene, Carlsbad, CA). Moreover, the DNase incubation was omitted, and DNA purified with the PowerClean DNA Clean-Up Kit (MOBIO laboratories, Carlsbad, CA) instead of the RNeasy Mini Kit (Qiagen, Valencia, CA).
PCR primers
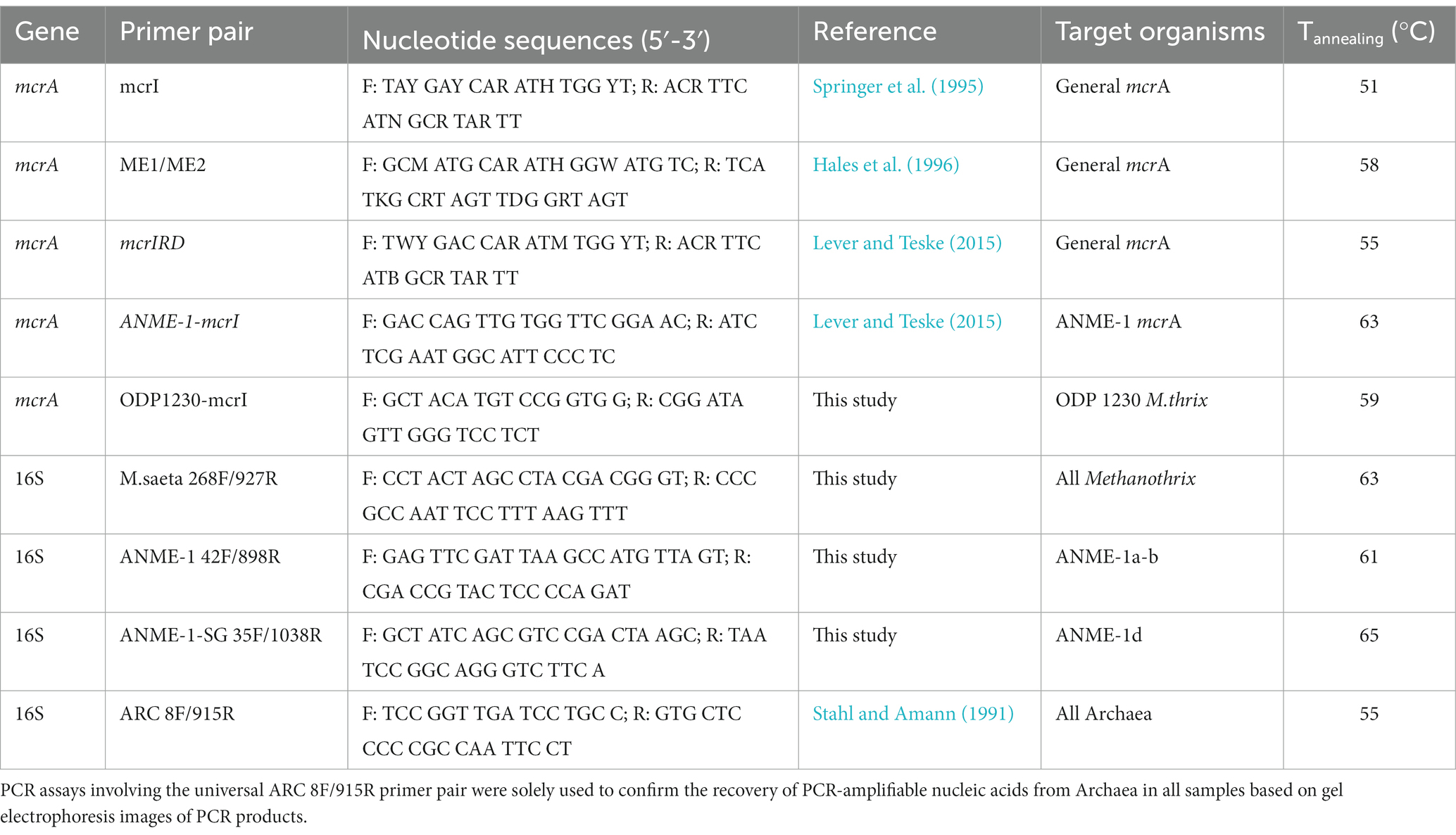
Two previously published general mcrA primer pairs yielded no amplification (ME1/ME2, Hales et al., 1996), or amplification at only one depth interval (mcrI, Springer et al., 1995; see Inagaki et al., 2006). The mcrIRD primer pair, a modified version of the mcrI primer pair with a reduced number of nucleotide degeneracies and consequently improved detection sensitivity, was used in conjunction with the ANME-1-specific ANME-1-mcrI primer pair [both published in Lever and Teske (2015)]. Special primers for ANME-1 detection were necessary due to the high number of nucleotide mismatches between the mcrI primer pair primer and the genetically divergent mcrA sequences of ANME-1. To confirm that detected mcrA detected belonged to active and living members of the methane-cycling community, we performed RT-PCR of mcrA-mRNA and 16S rRNA in several depth horizons using new group-specific primers for maximum amplification efficiency and hence detection sensitivity (mRNA: ODP1230, ANME-1; 16S rRNA: Msaeta 268F/927R, ANME-1 42F/898R; ANME-1-SG 35F/1038R). All primer sequences used in this study are shown in Table 1. All nucleotide sequences are publicly accessible at GenBank.
PCR protocols
PCR assays of mcrA were performed using the Takara SpeedSTAR HS DNA polymerase kit (TaKaRa Bio USA, Madison, WI) using (1) 1 × 2 min denaturation (98°C), (2) 40 × (a) 10s denaturation (98°C), (b) 30s annealing (Table 1 for temperatures), (c) 1 min extension (72°C), and (3) 1 × 5 min extension (72°C). Negative controls and reaction blanks were included.
RT-PCR assays were carried out using TaKaRa RNA PCR Kits (AMV) Version 3.0 (TaKaRa Bio USA, Madison, WI) and (1) 1 × 15 min reverse transcription, (2) 5 min denaturation (98°C), (3) 40 × (a) 30s denaturation (98°C), (b) 30s annealing (Table 1 for temperatures), (c) 1 min extension (72°C), and (4) 1 × 5 min extension (72°C). Negative controls and reaction blanks were included. Absence of DNA was confirmed by DNA-PCR with the same treatments but omitting the reverse transcription step.
Cloning and sequencing
PCR products were purified in a 2.5% low-melting point agarose gel using 1 × Tris acetate - EDTA buffer (TAE). Gel slices containing PCR fragments of the correct length were excised and purified using a S.N.A.P. Mini Kit (Invitrogen, Carlsbad, USA). Purified PCR fragments were cloned using the Topo TA Kit (Invitrogen, Carlsbad, USA) and transformed into TOP10 electrocompetent cells following the manufacturer’s instructions. Plasmid extraction and purification was done using the GeneJET Plasmid Miniprep Kit (ThermoFisher Scientific) and cycle sequencing was performed on an ABI 3730 Sequencer with M13 universal primers (SP010-SP030) at the Josephine Bay Paul Center at MBL (Woods Hole, MA). Sequences were BLAST analyzed using the nucleotide collection in GenBank.1 Phylogenetic trees were created and bootstrap analyses (1,000 replicates) performed in ARB2 using manually optimized SILVA 16S rRNA gene alignments, and a custom-built, publicly accessible mcrA database (name: mcrA4All)3 with >2,400 high-quality, aligned mcrA amplicon and genome sequences.
Thermodynamic calculations
Gibbs energy yields (ΔGr) of methanogenesis reactions from H2 + CO2 (2 HCO3− + 4 H2 + H+ ➔ CH4 + 3 H2O) and acetate (CH3COO− + H2O ➔ CH4 + HCO3−), and the methanogenic conversion of formate to methane [4 HCOO− + H2O + H+ ➔ CH4 + 3 HCO3−; note: this reaction presumably involves the initial oxidation of formate to H2 and HCO3−, which is not known to conserve energy except in certain hyperthermophiles (Schink et al., 2017)] were calculated based on the equation:
Results
Porewater gradients of chemical species determined on ODP Leg 201 provided the initial framework for our study and indicated ODP Site 1230 as a deep-sea site with unusually organic-rich sediments and highly active anaerobic microbial communities. Organic matter remineralization by microbes to at least 140 mbsf was indicated by DIC concentrations that increased steeply in the upper 25 mbsf and continued to increase gradually to 140 mbsf (Figure 2A). Sulfate reducing microbial communities depleted sulfate at ~9 mbsf in borehole A and up to 1 m deeper in boreholes B and C (Figure 2B). Porewater methane concentrations in borehole A were at background values (0.06 mM) at 6.10 mbsf, but had increased to 1.86 mM at 9.1 mbsf (Figure 2C). We thus estimate that the sulfate–methane transition zone (SMTZ), where most AOM takes place, was located within the depth interval from 7 to 9 mbsf in borehole A and up to 1 m deeper in boreholes B and C (Figure 2B, insert; for enlarged view of sulfate and methane profiles across the SMTZ in borehole A, see Supplementary Figure S1). Below the SMTZ, methane concentrations increased steeply, reaching saturation by ~28 mbsf, and hydrates appeared by ~50 mbsf. Hydrogen (H2) concentrations fluctuated greatly, but generally increased throughout the sulfate reduction zone, stabilized in the methanogenesis zone to ~140 mbsf, and decreased below (Figure 2D). Formate concentrations showed no clear depth-related trend and fluctuated between 3–15 μM throughout the entire cored interval (Figure 2E). By contrast, acetate concentrations increased from 3–11 μM in the sulfate reduction zone and SMTZ (upper 10 mbsf) to concentrations of ~20–60 μM in the methanogenesis zone between 30 to 140 mbsf (Figure 2E; see Supplementary Figure S2 for enlarged view of upper 20 mbsf). Below 140 mbsf, acetate concentrations rose sharply to 220 μM and remained >50 μM to the deepest cores sampled.
Carbon isotope geochemistry
13C-isotopic signatures of porewater methane and DIC provide insights into the zones of biological methane production and oxidation (Figure 3). Throughout the sediment column, methane was 13C-depleted relative to DIC. δ13C-DIC-values (only determined in borehole A) decreased slightly from −10.4‰ in the upper meter to −13.3‰ by 7.65 mbsf, then increased sharply in the uppermost methanogenic layer to +6‰ at 20 mbsf. The steepest increase in δ13C-DIC occurred within the interval from 7.65 mbsf (δ13C-DIC: −13.2‰) to 9.15 mbsf (δ13C-DIC: −8.4‰), and suggests onset of methanogenesis by CO2 reduction in this interval. Below 20 mbsf, δ13C-DIC-values continued to gradually increase to reach a maximum of +20‰ at 123 mbsf, below which values slightly fell off to +15‰ at 246 mbsf (Figure 3). The 13C-isotopic compositions of methane, determined in the methanogenesis zones of boreholes A (>25 mbsf) and B (0–12 mbsf), were in a range typical of biological methanogenesis (Whiticar et al., 1986; Whiticar, 1999). δ13C-CH4 was ~ −65‰ in the upper 6 mbsf, and then decreased to −75‰ at 12 mbsf. This increase in δ13C-CH4 upward through the SMTZ is consistent with isotopic discrimination of AOM against 13C-CH4. Below, values gradually increased from to ~ −65‰ at 246 mbsf (Figure 3). The difference in δ13C-CH4 relative to δ13C-DIC was ~ −53‰ in the upper 6 mbsf, decreased across the SMTZ reaching −71‰ in the upper methanogenesis zone at 12 mbsf, and stabilized at −80‰ to −85‰ below 25 mbsf (Figure 3).
mcrA sequence diversity
We detected mcrA sequences of three phylogenetic groups (Figure 4): (1) putatively anaerobic methanotrophic ANME-1a-b Archaea, (2) a sister group of ANME-1, which we here refer to as ANME-1d, and (3) sequences of Methanotrichales that cluster with a genus-level group that includes the known aceticlastic methanogens Methanothrix harundinacea and Methanothrix pelagica.
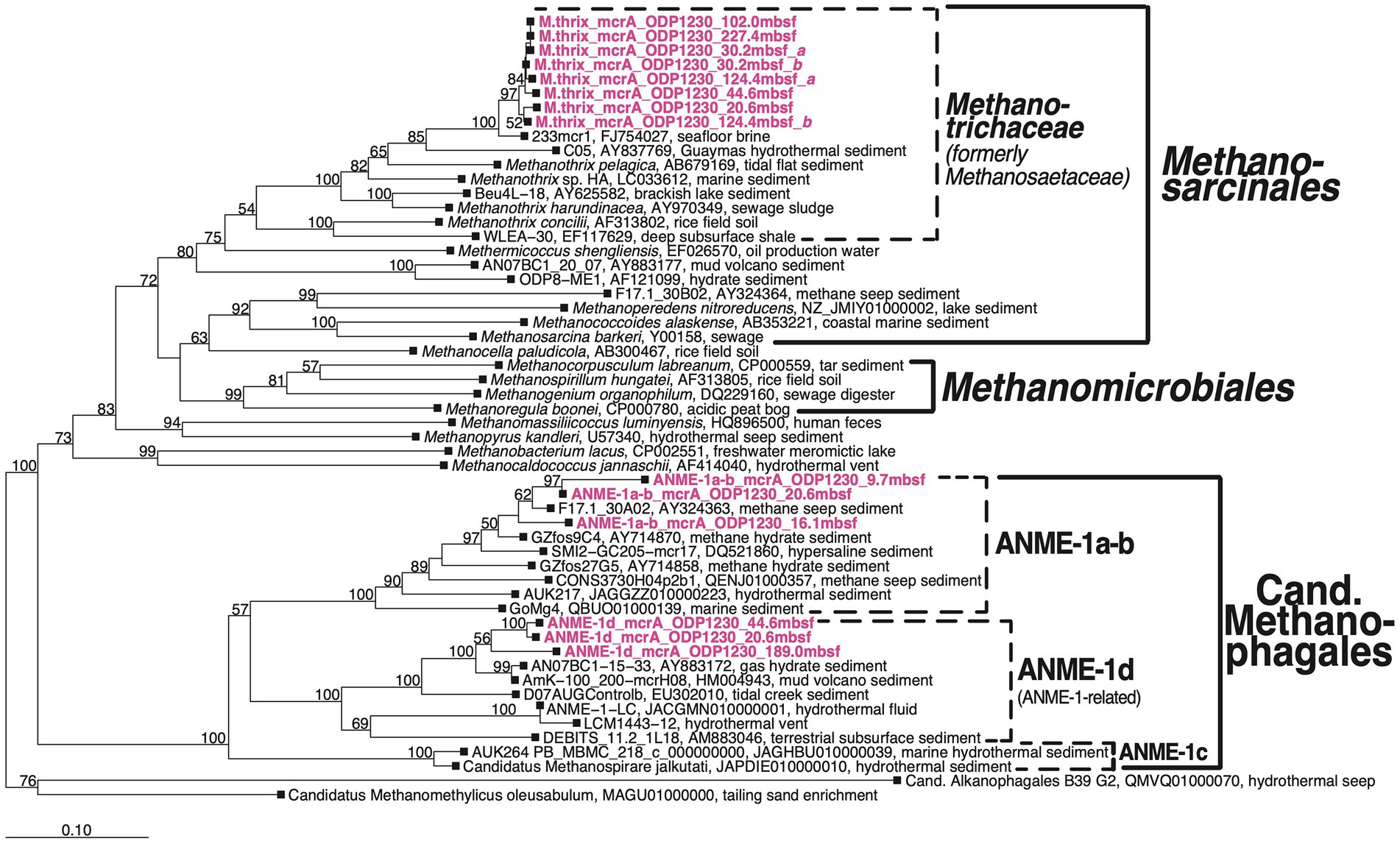
Figure 4. Bootstrap phylogenetic tree based on mcrA nucleotide sequences. Created using Jukes-Cantor correction in ARB neighbor-joining (Ludwig et al., 2004). Cand., Candidatus.
The three groups were vertically zonated (Figure 5). ANME-1a-b mcrA sequences were found in horizons near the upper (7.8 mbsf) and lower limit (9.7 mbsf) of the STMZ in Borehole B and four horizons in the upper methanogenesis zone (10.25–20.6 mbsf; Table 2). Sequences of ANME-1d were detected with ANME-1a-b sequences at one depth in the upper methanogenesis zone (20.6 mbsf) and in three horizons below (to 189.0 mbsf). Methanothrix mcrA showed a distribution similar to ANME-1d, but was detected in more sediment horizons and to greater depth (to 227 mbsf; Table 2).
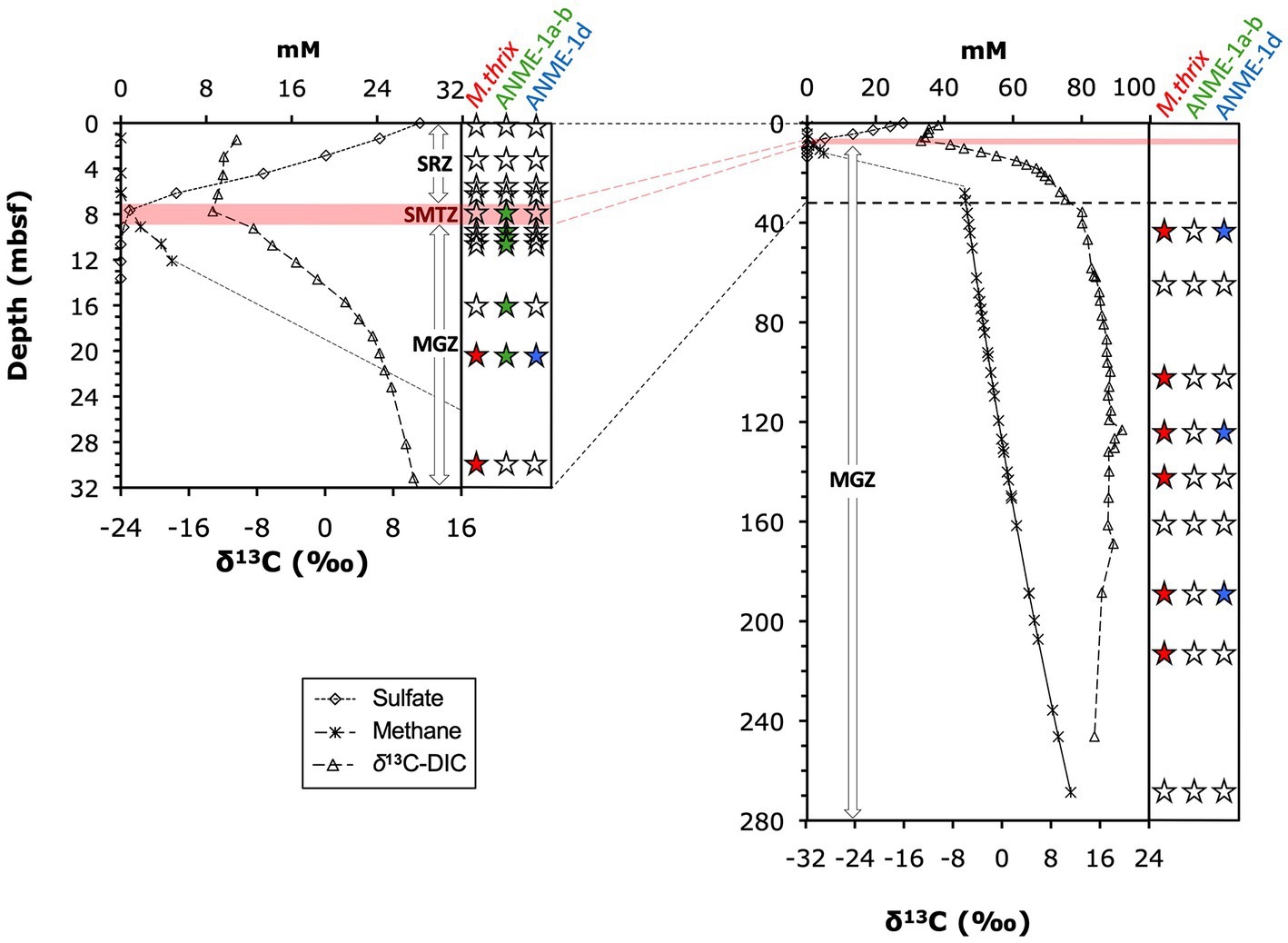
Figure 5. Distribution of mcrA groups along depth and geochemical gradients of sulfate, methane, and δ13C-DIC. Panel on right side of each graph indicates detection/absence of detection of (1) ANME-1a-b, (2) ANME-1d, and (3) Methanothrix sequences. Solid black symbols indicate detection, empty symbols indicate lack of detection (example:  indicates presence of ANME-1, and absence of ANME-1d and Methanothrix). Horizontal red bar indicates the depth interval of the SMTZ, where most AOM takes place, for Borehole A (7 to 9 mbsf). This interval extended ~1 m deeper (~10 mbsf) in Borehole B. The uppermost detections of ANME1-a-b mcrA were at 7.8 and 9.7 mbsf in Borehole B and thus near the upper and lower limits of the SMTZ in this borehole.
indicates presence of ANME-1, and absence of ANME-1d and Methanothrix). Horizontal red bar indicates the depth interval of the SMTZ, where most AOM takes place, for Borehole A (7 to 9 mbsf). This interval extended ~1 m deeper (~10 mbsf) in Borehole B. The uppermost detections of ANME1-a-b mcrA were at 7.8 and 9.7 mbsf in Borehole B and thus near the upper and lower limits of the SMTZ in this borehole.
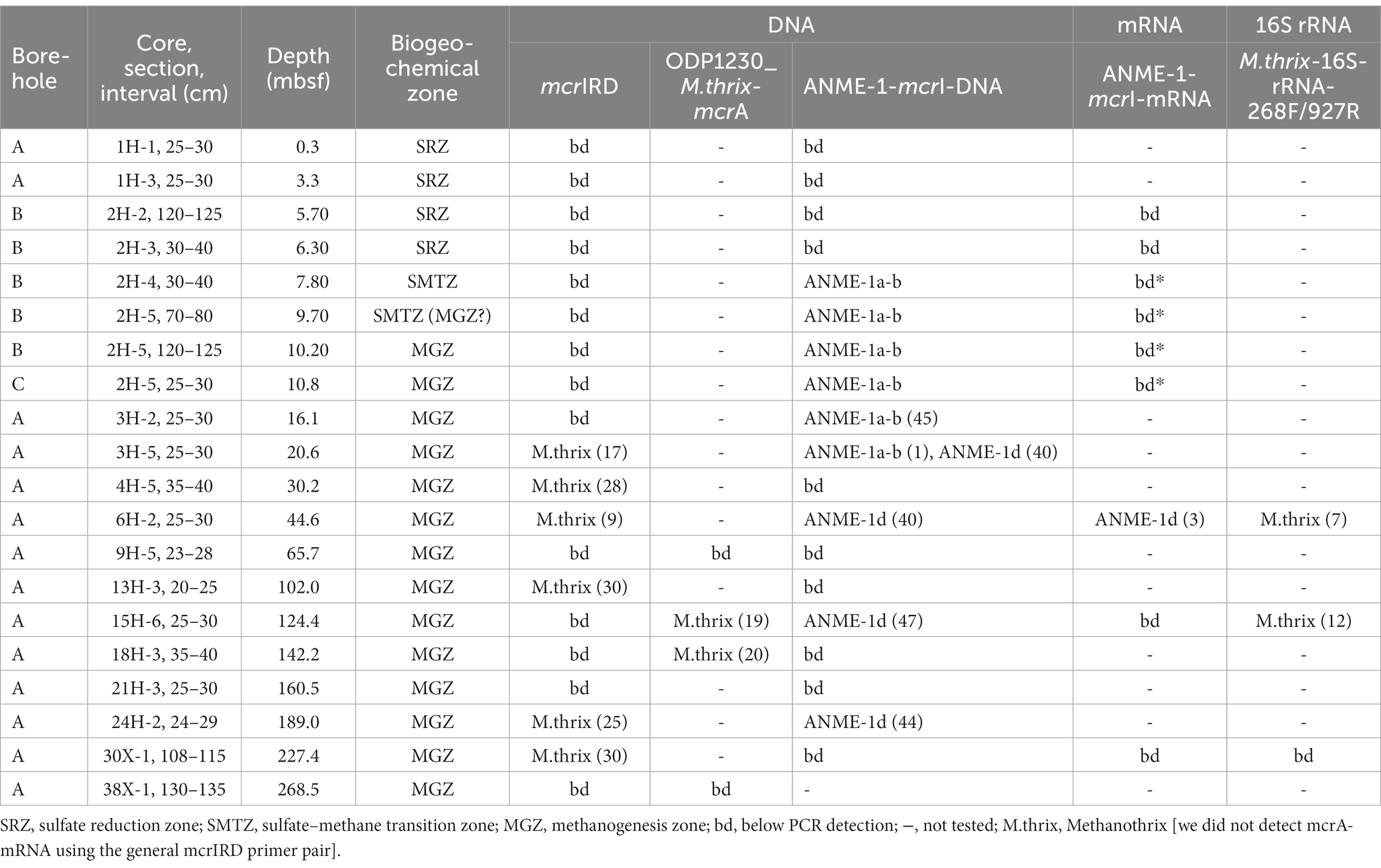
Table 2. Overview of boreholes, core samples, sediment depths, and biogeochemical zones from which DNA and RNA were extracted, and the results of PCR amplifications with different primers with number of clones sequenced in parentheses.
We detected mcrA-mRNA of ANME-1-d in one of the 9 samples examined using the ANME-1-mcrA primer pair (core 6H-2, 44 mbsf; Table 2). In 11 replicate RT-PCRs of RNA extracts, controls (PCR negative, extraction blank, DNA controls) always tested negative, whereas 8/11 RNA extracts tested positive. By comparison, four other samples (from 7.8, 9.7, 10.2, 10.8 mbsf) yielded RT-PCR detection with the same primers, but DNA controls were positive (albeit weaker than cDNA bands), indicating that traces of DNA had resisted the DNAse treatment. We cloned cDNA of mcrA-mRNA from core 6H-2 and confirmed the presence of mcrA of ANME-1d.
In addition to mcrA-mRNA, we examined 16S rRNA sequences. RT-PCRs with new Methanothrix-specific 16S rRNA gene primers (Table 1) yielded Methanothrix-like sequences in two additional depth horizons (Table 2; Supplementary Figure S3). This primer pair also generated 16S rRNA sequences of a sister group of Methanosarcinales, previously detected in methane seep and mud volcano environments, with unknown metabolism at two depths (15H-6, 30H-1; Supplementary Figure S3). Interestingly, despite detecting mRNA with ANME-1-mcrA primers, we were unable to detect 16S rRNA of ANME-1a-b or ANME-1d with newly designed group-specific 16S rRNA gene primers (Table 1).
Discussion
We present a depth profile of mcrA that relates distribution patterns of deep subseafloor methanogens and anaerobic methanotrophs to the geochemical context. While the genetic and gene transcript analyses in our study are present-day snapshots of methane-cycling activity, the measured geochemical data in part capture much longer time scales, such as the accumulation of methane over millions of years. Nonetheless, we observe a clear relationship between the community profile of methane-cycling archaea and porewater geochemical gradients. We, moreover, resolve the paradox of earlier studies in which methane-rich sediments at ODP Site 1230 appeared largely devoid of methanogens in the methanogenesis zone (Inagaki et al., 2006), and completely devoid of anaerobic methanotrophs in the SMTZ (Biddle et al., 2006).
Geochemical and functional gene profiles indicate a distinct depth stratification of the active methane cycling community (Table 2; Figure 5). No nucleic acid evidence of present-day methane-cycling was detected in the upper part of the sulfate reduction zone (0 to ~7 mbsf) despite methane concentrations in the micromolar range. Throughout the SMTZ (~7 to 9 mbsf in borehole A, up to 1 m deeper in boreholes B and C), sulfate concentrations diminished in typical concave-down profiles, and methane concentrations increased. These gradients coincide with the detection of mcrA of ANME-1a-b (Figure 5), members of which are known to be anaerobic methanotrophs (Knittel and Boetius, 2009). Therefore, our sulfate and methane concentration profiles and mcrA composition in the SMTZ are consistent with AOM. In the underlying methanogenesis zone (~9 to 269+ mbsf), methane concentrations and δ13C-DIC increase drastically in the upper tens of meters and stay high throughout, while sulfate remains depleted (Figures 2, 3). Interestingly, ANME-1a-b Archaea were detected in the upper meters of the methanogenesis zone (from 9.7 to 16.1 mbsf), in line with past indications that ANME-1a-b might be capable of methanogenesis in addition to methanotrophy (House et al., 2009; Lloyd et al., 2011; Beulig et al., 2019). Moreover, ANME-1a-b were vertically separated from ANME-1d and Methanothrix mcrA sequences, which were only found in deeper, methanogenic sediment layers. The three groups only overlapped in core 3H-5 (20.6 mbsf), which marked the deepest sample in which ANME-1a-b and shallowest sample in which ANME-1d and Methanothrix were detected.
Implications of the 13C-isotopic data
The changes in δ13C-DIC and –methane provide insights into the sources of DIC and pathways of methane production at ODP Site 1230. The δ13C-DIC isotopic values in the upper ~9 mbsf (−10.4 to −13.2‰) are consistent with organic matter mineralization becoming the main DIC source with increasing sediment depth. Most of this organic matter is likely to be phytoplankton-derived organic matter (δ13C-total organic carbon: ~22–23‰; Biddle et al., 2006) that was initially deposited under the upwelling regime of the Peru Margin, and subsequently reworked and laterally transported downslope to the Peru Trench. Toward the sediment surface, the δ13C-DIC increases, most likely due to an increasing contribution of 13C-enriched DIC from deep sea bottom water, which typically bears a 13C-composition of ~0 to +1.2‰ (Lynch-Stieglitz et al., 1995).
Notably, despite the strong geochemical evidence for AOM in the SMTZ, which might be expected to produce highly 13C-depleted DIC from the oxidation of methane, we do not observe a strong downward swing in δ13C-DIC within the SMTZ. This phenomenon has been observed previously in SMTZs and has been explained with concomitant AOM and methane production (Beulig et al., 2019), microbially mediated isotope exchange between methane and DIC (Yoshinaga et al., 2014), and reversibility of intracellular methane-cycling reactions at low sulfate concentrations (Wegener et al., 2021). In our case, the mcrA data argue against the first scenario, if ANME-1a-b are assumed to only perform methanotrophy. Yet, if – as proposed previously - ANME-1a-b are facultative methanogens, which matches the detection of this group throughout the upper ~12 m of the methanogenesis zone, then the first scenario is also plausible. Notably, porewater dissolved barium concentrations increase sharply throughout the AOM and upper methanogenesis zone (e.g., from 2.7 μM at 6.15 mbsf to 290 μM at 23.15 mbsf at ODP Site 1230A; D’Hondt et al., 2003), consistent with (slow) release of sulfate through chemical dissolution of barite (BaSO4). This sulfate could fuel low rates of AOM, and thus also support concomitant AOM and methane production throughout the upper methanogenesis zone. AOM coupled to iron or manganese reduction could also support low rates of AOM, as was recently proposed for subsurface sediments of the South China Sea, where ANME-1 were detected meters below the SMTZ (Zhang et al., 2023). Yet, the low porewater concentrations of Fe2+ (0.6 to 3.9 μM) and Mn2+(0 to 0.3 μM) in the upper methanogenic sediment layer where we detected ANME-1a-b at ODP Site 1230 (D’Hondt et al., 2003) do not support an important role of AOM coupled to metal reduction.
Below 9 mbsf, the δ13C-DIC increased, consistent with a strong isotopic imprint of methanogenesis by CO2 reduction. Strong isotopic discrimination against 13C-CO2 is the norm in methanogenesis from H2/CO2 (Whiticar, 1999; Penning et al., 2005) and can result in significant 13C-enrichment of the residual DIC pool (Alperin and Hoehler, 2009; House et al., 2009). Based on measured porewater geochemical data, methanogenesis from H2/CO2 is, however, not thermodynamically favorable (Figure 6), with in situ Gibbs energies in the positive (i.e., endergonic) range (ΔG r ’ > 0 kJ mol−1) throughout the sediment column of ODP Site 1230. Since methanogenesis from formate follows the same biochemical route as hydrogenotrophic methanogenesis after the initial oxidation of formate to CO2 and H2 by formate dehydrogenase (Sparling and Daniels, 1986), a similar isotopic fractionation can be expected. Indeed, the complete conversion reaction of formate to methane is thermodynamically favorable, and based on that alone formate a potential methanogenic substrate at ODP Site 1230 (Figure 6). Yet, assuming that energy is not conserved during the initial formate oxidation step, but only in the second step involving methanogenic CO2 reduction with H2 (Schink et al., 2017), then formate conversion to methane appears less plausible. This is because intracellular H2 concentrations can be expected to be close to equilibrium with H2 concentrations in the surrounding sediment due to H2 leakage out of methanogenic cells (Finke et al., 2007). As stated above, however, measured H2 concentrations in the surrounding sediment are too low to energetically support hydrogenotrophic methanogenesis. A more recently documented form of methanogenic CO2 reduction involves interspecies electron transfer (IET). This form of methanogenesis, which was first discovered in Methanothrix harundinaceae (Rotaru et al., 2014), involves cellular structures, e.g., cytochromes, that attach to conductive mineral surfaces or syntrophic partner organisms (Gao and Lu, 2021). The isotopic fractionations of these reactions are not known but most likely also cause δ13C-enrichment of residual DIC. In principle, the conversion of formate to methane could also operate via a direct electron transfer mechanism, e.g., from syntrophic bacteria to methanogens. This mechanism could bypass H2 as a catabolic intermediate and even render formate catabolism a potential source of methane. Thus, based on the available geochemical data, the dominance of methanogenic CO2 reduction at Site 1230, which was inferred from the δ13C-DIC profile below 9 mbsf, is most plausibly explained with electron transfer from syntrophic partner organisms or mineral surfaces to methanogens.
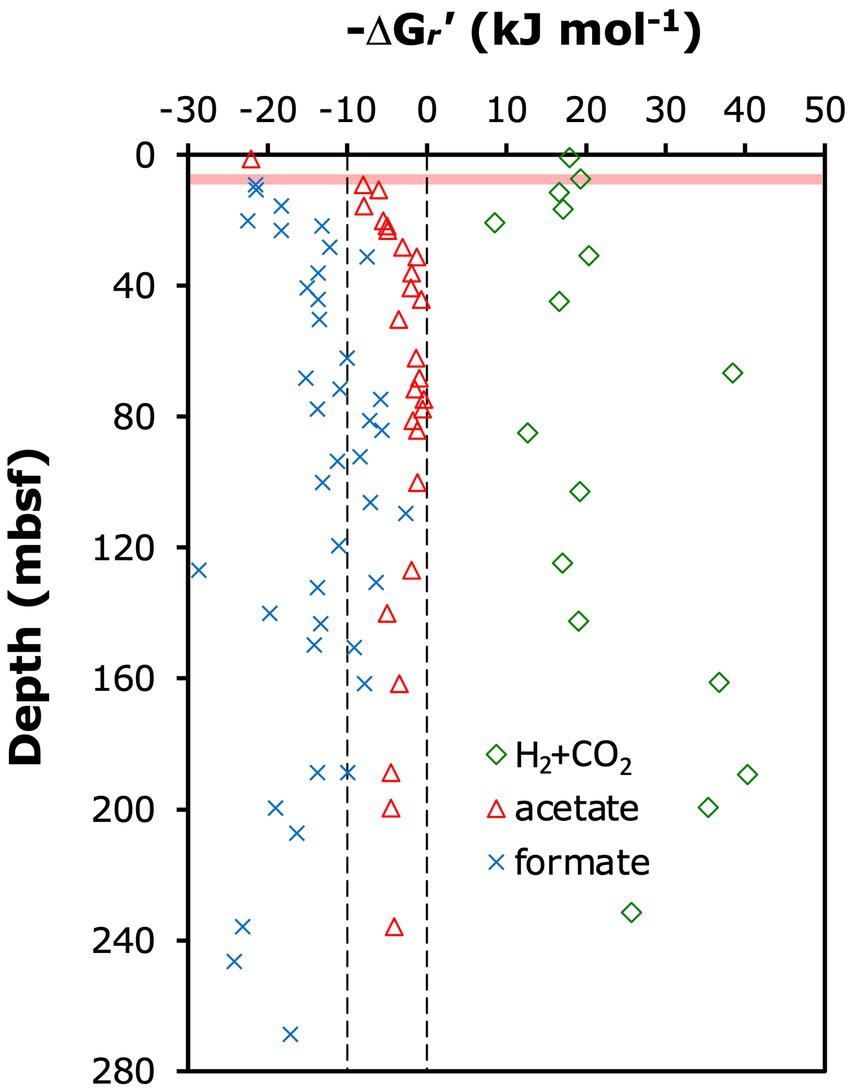
Figure 6. Calculated in situ Gibbs energy yields (ΔGr’) of methanogenesis reactions from H2 + CO2 (HCO3– + 4 H2 + H+ ➔ CH4 + 3 H2O) and acetate (CH3COO– + H2O ➔ CH4 + HCO3–), and the methanogenic conversion of formate to methane (4 HCOO– + H2O + H+ ➔ CH4 + 3 HCO3–; note: this reaction includes the initial oxidation of formate to H2 and HCO3–, which may not be coupled to energy conservation). Calculations were done for in situ conditions as described in the Materials & Methods.
By contrast, chemoautotrophy, acetogenesis or other methanogenic pathways are unlikely drivers of the observed δ13C-DIC increase in the methanogenesis zone. Although (certain) ANME-1a-b are chemoautotrophs (Kellermann et al., 2012), past studies indicate that AOM of isotopically highly depleted methane (~ − 75 per mil) to CO2 occurs at much (≥40-fold) higher rates than C-assimilation by chemoautotrophy (e.g., Nauhaus et al., 2007; Wegener et al., 2008). Consequently, AOM would be expected to overprint any C-isotopic enrichment of DIC by chemoautotrophy. Acetogenesis from H2/CO2, which also strongly discriminates against 13C (Gelwicks et al., 1989), is unlikely based on δ13C-acetate values of −12 to −18‰ at ODP Site 1230 that indicate fermentation as the main acetate source (Heuer et al., 2006). The other widespread methanogenesis pathways from acetate (aceticlastic methanogenesis) and methylated substrates (e.g., methanol, dimethyl sulfide and methyl amines; methylotrophic methanogenesis), produce rather than consume CO2 (Whitman et al., 2014). In aceticlastic methanogenesis, this CO2 has the same 13C-depleted isotopic composition as the methane produced (Gelwicks et al., 1994) and would thus lower (rather than increase) the δ13C-DIC. Despite high concentrations of acetate, our calculations, moreover, indicate that aceticlastic methanogenesis is close to thermodynamic equilibrium throughout most of the methanogenesis zone (Figure 6), with Gibbs energies not reaching the theoretical minimum required for biological energy conservation by proton translocation (ΔG r ’ ≅ 10 kJ mol−1; Hoehler et al., 2001; Lever et al., 2015). 13C-depletion of DIC is also expected for methylotrophic methanogenesis, even though this pathway produces methane with similar isotopic fractionations as CO2 reduction (Conrad, 2005). The reason for 13C-depletion of DIC is that the main isotopic fractionation of methylotrophic methanogenesis is produced by the first enzymes in the reaction chain (methyl transferase I and/or II; Krzycki et al., 1987) and hence upstream of where C fractions enter separate enzymatic pathways to produce CO2 and methane (Thauer, 1998). Notably, another form of methylotrophic methanogenesis, which involves methylated substrates and hydrogen, e.g., methanol + H2 does not produce CO2 (Dridi et al., 2012; Whitman et al., 2014). A fourth methanogenic pathway that involves the conversion of methoxy-groups from lignin monomers to methane with CO2 as a co-substrate (Mayumi et al., 2016) is in theory also possible. Yet, this pathway is unlikely to be important given the primarily phytoplanktonic origin of organic matter at ODP Site 1230 (Shipboard Scientific Party, 1988; D’Hondt et al., 2003) and recent evidence suggesting minimal long-term degradation of lignin in anoxic sediment (Han et al., 2022).
The increase in δ13C-DIC with depth is steepest from the lower SMTZ to ~20 mbsf (Figures 3, 5), consistent with rates of methanogenic CO2 reduction being highest in this interval. The subsequent decrease in the slope of δ13C-DIC with depth can be explained with an increase in the DIC pool size and decline in the rates of CO2 reduction. In addition, it is possible that the relative contributions of other methanogenic pathways, e.g., aceticlastic methanogenesis, increase below this depth. Nonetheless, the 13C-isotopic depletions of −80 to −86‰ of δ13C-methane relative to δ13C-DIC that were consistently measured below 18 mbsf (Figure 3) indicate that CO2 reduction accounts for most of the methane that has accumulated throughout the methanogenesis zone of ODP Site 1230.
Community zonation
When examined in the geochemical context, the distribution of mcrA genes within the methanogenesis zone of ODP Site 1230 may be surprising. Isotopic compositions of DIC and methane suggest predominance of methanogenesis by CO2 reduction, whereas detected mcrA sequences belong to phylotypes of putatively anaerobic methane-oxidizing ANME-1a-b, its catabolically uncharacterized sister group ANME-1d, and Methanothrix, a genus that was traditionally believed to consist uniformly of obligately aceticlastic methanogens.
The presence of ANME-1a-b in the SMTZ and in underlying sediment that is net methanogenic can be explained with different scenarios. The first one is that ANME-1a-b are indeed facultatively methanogenic, as proposed previously based on strong heterogeneity in δ13C of ANME-1-biomass in seep sediments (House et al., 2009), detection of ANME-1a-b mcrA transcripts in methanogenic sediment (Lloyd et al., 2011), and combined methanogenesis rate measurements and mcrA analyses across anaerobic methane-oxidizing and methanogenic sediment (Beulig et al., 2019). This environmental evidence has been supported by recent genomic detections of hydrogenase genes, that are potentially involved in hydrogenotrophic methanogenesis, across multiple ANME-1a-b and ANME-1c taxa (Laso-Pérez et al., 2023). Alternatively, AOM by ANME-1a-b may continue as a cryptic process in the presence of methanogenesis throughout the uppermost part of the methanogenesis zone. Our thermodynamic calculations indicate that a reversal of methanogenic CO2 reduction with H2 is thermodynamically favorable throughout bulk sediments of ODP Site 1230 (Figure 6), though the electron acceptor is unclear. As discussed earlier, the increase in dissolved barium indicates barite (BaSO4) dissolution in this part of the sediment column as a potential source of sulfate, whereas the very low Fe2+ and Mn2+ concentrations argue against a significant role of AOM coupled to metal reduction. Under this scenario, the organisms that were responsible for the production of the measured methane are unknown. While neither possibility can be ruled out, the available evidence from this and past studies supports ANME-1a-b contributing to the production of methane by CO2 reduction in the upper methanogenesis zone of ODP 1230.
The metabolism of ANME-1d, which replaces ANME-1a-b in deeper sediment layers of the methanogenesis zone, is even less understood than that of ANME-1a-b. This group, which was previously also referred to as “ANME-1-related group” and represents a poorly studied, sister family, or even sister order, of ANME-1a-b (Lever and Teske, 2015), has been found across a range of anoxic environments. These include hydrothermal vents in ultramafic settings (Kelley et al., 2005), deeply buried terrestrial coalbeds (Fry et al., 2009), marine gas hydrate sediments (Kormas et al., 2005), and tidal creek sediments (Edmonds et al., 2008). Given the sole detection of ANME-1d DNA and mRNA deep in the methanogenesis zone, a methanogenic lifestyle seems likely. This group could reduce CO2 to methane using electrons from IET and thus contribute to the observed strong δ13C-depletion of methane relative to DIC.
The predominant detection of Methanothrix-mcrA sequences below 20 mbsf, despite δ13C-DIC compositions that indicate mainly methanogenesis by CO2 reduction, and Gibbs energies of aceticlastic methanogenesis near thermodynamic equilibrium, is perplexing, given that members of Methanothrix are traditionally considered to be obligate aceticlasts. One explanation is that these sequences belong to inactive or dead cells. Yet, this explanation does not match the detection of rRNA of Methanothrix (Table 2), and is at odds with research suggesting that the vast majority of DNA from dead microorganisms is degraded over time scales of centuries in subsurface sediments (Torti et al., 2018). Instead, the Methanothrix mcrA and 16S rRNA sequences may not belong to (obligate) aceticlasts. While genomic data of Methanothrix thermophila indicate potential for hydrogenotrophic metabolism in this group (Smith and Ingram-Smith, 2007), methanogenesis involving H2 has never been shown for Methanothricaceae. Yet, more recent experiments with pure cultures have demonstrated that members of Methanothrix - including Methanothrix harundinacea, which mcrA sequences from ODP Site 1230 cluster with (Figure 5) - are capable of methanogenic growth by CO2 reduction using electrons received directly or through mineral intermediates from syntrophic partner organisms (Rotaru et al., 2014; Yang et al., 2019; Gao and Lu, 2021). Experiments involving rice paddy soils and lake sediments have provided additional evidence for CO2 reduction by Methanothrix in the environment (Holmes et al., 2017; Rotaru et al., 2019). Consequently, the observed δ13C-DIC and δ13C-methane compositions and dominance of mcrA sequences of Methanothrix may not be a contradiction, but instead match revised knowledge on the metabolic capabilities of Methanothrix.
Conclusion
We provide the first complete community profile of active methane-cycling archaea in deep subseafloor sediments, and show based on DNA and RNA sequence data that anaerobic methane-cycling archaea are present throughout the SMTZ and methanogenesis zone of ODP Site 1230 in the Peru Trench. Of essential importance for the detection of mcrA, mcrA-mRNA, and 16S rRNA of methane-cycling archaea was the use of redesigned general mcrA primers and development of new group-specific mcrA and 16S rRNA gene primers. While these primers improved the detection sensitivity of methane-cycling archaea, they confirm the notion that methane-cycling archaea only account for a small fraction of deep subsurface microbial communities, even in AOM and methanogenesis zones (Lever, 2013).
Even though DNA- and RNA-based detections of methane-cycling archaea generally match the distributions of AOM and methanogenesis based on geochemical data, the detected phylogenetic groups appear at odds with the inferred dominant methane-cycling pathways. ANME-1, which are historically considered to be anaerobic methanotrophs, were detected to sediment depths that were > 10 m (ANME-1a-b) and > 100 m (ANME-1d) below the SMTZ. Based on published sedimentation rates for Site 1230 (0.25 mm yr.−1; Shipboard Scientific Party, 1988), these distances suggest the continued existence of ANME-1a-b and ANME-1d populations in methanogenic sediments for >40,000 and > 400,000 years after their burial below the SMTZ, respectively. Given the measured methane concentration and DIC-isotopic data, and that no other methane-cycling archaea were detected, a switch to methanogenesis by CO2 reduction offers the most parsimonious explanation for the occurrence of ANME-1a-b far below the SMTZ. Similarly, methanogenesis by CO2 reduction may sustain populations of ANME-1d in deeper layers, and also explain why members of Methanothrix – that were historically assumed to be aceticlastic - are pervasive throughout sediments that appear to be dominated by methanogenic CO2 reduction. Herein, the pathway of CO2 reduction remains unclear, but could bypass H2 as an electron source through direct electron transfer.
Data availability statement
The original contributions presented in the study are publicly available. This data can be found here: All isotopic data (d13C-CH4, d13C-DIC) are included in Supplementary Table 1. All geochemical concentration data, including pH, are publicly available in D’Hondt et al. (2003). All nucleotide sequences can be retrieved from GenBank (mcrA: OQ603043-OQ603056; 16S rRNA: OQ658172-OQ658186).
Author contributions
ML, MA, and AT designed the research. AT and K-UH obtained the samples. ML and K-UH produced the data. ML analyzed the data with input from MA and AT and wrote the manuscript with input from all co-authors. All authors contributed to the article and approved the submitted version.
Funding
Sequencing was supported by the NASA Astrobiology Institute “From Early Biospheric Metabolisms to the Evolution of complex systems” and performed at the Josephine Bay Paul Center for Comparative Molecular Biology and Evolution at the Marine Biological Laboratory, Woods Hole, MA. ML was supported by a Schlanger Ocean Drilling Fellowship, and a University of North Carolina Dissertation Completion Fellowship.
Acknowledgments
The authors thank the Ocean Drilling Program (ODP) and in particular the ODP Leg 201 Shipboard Scientific Party for sampling support, Ketil Sørensen and Christopher S. Martens for helpful discussions, and Barbara J. MacGregor for intellectual input to earlier manuscript versions.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1192029/full#supplementary-material
Footnotes
1. ^www.ncbi.nlm.nih.gov/blast
3. ^https://drive.google.com/drive/u/0/folders/1G8GeJuYsIX4MLv5-LaUQHD9f9F9rfIAu
References
Alperin, M. J., and Hoehler, T. M. (2009). Anaerobic methane oxidation by Archaea/sulfate-reducing bacteria aggregates: 2 Isotopic constraints. Am. J. Sci. 309, 958–984. doi: 10.2475/10.2009.02
Beulig, F., Røy, H., McGlynn, S. E., and Jørgensen, B. B. (2019). Cryptic CH4 cycling in the sulfate-methane transition of marine sediments apparently mediated by ANME-1 archaea. ISME J. 13, 250–262. doi: 10.1038/s41396-018-0273-z
Biddle, J. F., Fitz-Gibbon, S., Schuster, S. C., Brenchley, J. E., and House, C. H. (2008). Metagenomic signatures of the Peru margin subseafloor biosphere show a genetically distinct environment. Proc. Natl. Acad. U. S. A. 105, 10583–10588. doi: 10.1073/pnas.0709942105
Biddle, J. F., Lipp, J. S., Lever, M. A., Lloyd, K., Sørensen, K., Anderson, R., et al. (2006). Heterotrophic Archaea dominate sedimentary subsurface ecosystems off Peru. Proc. Natl. Acad. U. S. A. 103, 3846–3851. doi: 10.1073/pnas.0600035103
Carr, S. A., Schubotz, F., Dunbar, R. B., Mills, C. T., Dias, R., Summons, R. E., et al. (2018). Acetoclastic Methanosaeta are dominant methanogens in organic-rich Antarctic marine sediments. ISME J. 12, 330–342. doi: 10.1038/ismej.2017.150
Colwell, F. S., Boyd, S., Delwiche, M. E., Reed, D. W., Phelps, T. J., and Newby, D. T. (2008). Estimates of biogenic methane production rates in deep marine sediments at hydrate ridge, Cascadia margin. Appl. Environ. Microbiol. 74, 3444–3452. doi: 10.1128/AEM.02114-07
Conrad, R. (2005). Quantification of methanogenic pathways using stable carbon isotopic signatures: a review and a proposal. Org. Geochem. 36, 739–752. doi: 10.1016/j.orggeochem.2004.09.006
Cragg, B. A., Parkes, R. J., Fry, J. C., Herbert, R. A., Wimpenny, J. W. T., and Getliff, J. M. (1990). “Bacterial biomass and activity profiles within deep sediment layers” in Proc. ODP, Scientific Res. eds. E. Suess and R. von Huene, vol. 112 (College Station, TX: Ocean Drilling Program)
D’Hondt, S. L., Jørgensen, B. B., and Miller, D. J.. (2003). 1. Leg 201 summary. Proc. ODP, Init. Repts. College Station, TX: Ocean Drilling Program. 201.
D’Hondt, S., Jørgensen, B. B., Miller, D. J., Batzke, A., Blake, R., Cragg, B. A., et al. (2004). Distributions of microbial activities in deep subseafloor sediments. Science 306, 2216–2221. doi: 10.1126/science.1101155
Deng, L., Bölsterli, D., Kristensen, E., Meile, C., Su, C.-C., Bernasconi, S. M., et al. (2020). Macrofaunal control of microbial community structure in continental margin sediments. Proc. Natl. Acad. Sci. U. S. A. 117, 15911–15922. doi: 10.1073/pnas.1917494117
Dridi, B., Fardeau, M.-L., Ollivier, B., Raoult, D., and Drancourt, M. (2012). Methanomassiliicoccus luminyensis gen. Nov., sp. nov., a methanogenic archaeon isolated from human faeces. Int. J. Syst. Evol. Microbiol. 62, 1902–1907. doi: 10.1099/ijs.0.033712-0
Edmonds, J. W., Weston, N. B., Joye, S. B., and Moran, M. A. (2008). Variation in prokaryotic community composition as a function of resource availability in tidal creek sediments. Appl. Environ. Microbiol. 74, 1836–1844. doi: 10.1128/AEM.00854-07
Finke, N., Hoehler, T. M., and Jørgensen, B. B. (2007). Hydrogen ‘leakage’ during methanogenesis from methanol and methylamine: implications for anaerobic carbon degradation pathways in aquatic sediments. Environ. Microbiol. 9, 1060–1071. doi: 10.1111/j.1462-2920.2007.01248.x
Friedrich, M. W. (2005). Methyl-coenzyme M reductase genes: unique functional markers of methanogenic and anaerobic methane-oxidizing Archaea. Meth. Enzymol. 397, 428–442. doi: 10.1016/S0076-6879(05)97026-2
Fry, J. C., Horsfield, B., Sykes, R., Cragg, B. A., Heywood, C., Kim, G. T., et al. (2009). Prokaryotic populations and activities in an interbedded coal deposit, including a previously deeply buried section (1.6–2.3 km) above ∼150 ma basement rock. Geomicrobiol J. 26, 163–178. doi: 10.1080/01490450902724832
Futagami, T., Morono, Y., Terada, T., Kaksonen, A. H., and Inagaki, F. (2009). Dehalogenation activities and distribution of reductive dehalogenase homologous genes in marine subsurface sediments. Appl. Environ. Microbiol. 75, 6905–6909. doi: 10.1128/AEM.01124-09
Gao, K., and Lu, Y. (2021). Putative extracellular electron transfer in methanogenic archaea. Front. Microbiol. 12:611739. doi: 10.3389/fmicb.2021.611739
Gelwicks, J. T., Risatti, J. B., and Hayes, J. M. (1989). Carbon isotope effects associated with autotrophic acetogenesis. Org. Geochem. 14, 441–446. doi: 10.1016/0146-6380(89)90009-0
Gelwicks, J. T., Risatti, J. B., and Hayes, J. M. (1994). Carbon isotope effects associated with aceticlastic methanogenesis. Appl. Environ. Microbiol. 60, 467–472. doi: 10.1128/aem.60.2.467-472.1994
Hales, B. A., Edwards, C., Ritchie, D. A., Hall, G., Pickup, R. W., and Saunders, J. R. (1996). Isolation and identification of methanogen-specific DNA from blanket bog peat by PCR amplification and sequence analysis. Appl. Environ. Microbiol. 62, 668–675. doi: 10.1128/aem.62.2.668-675.1996
Han, X., Tolu, J., Deng, L., Fiskal, A., Schubert, C. J., Winkel, L. H. E., et al. (2022). Physical shielding promotes long-term preservation of biomolecules in lake sediments. PNAS Nexus 1, 1–15. doi: 10.1093/pnasnexus/pgac076
Heuer, V. B., Elvert, M., Tille, S., Krummen, M., Prieto Mollar, X., Hmelo, L. R., et al. (2006). Online δ13C analysis of volatile fatty acids in sediment/porewater systems by liquid chromatography-isotope ratio mass spectrometry. Limnol. Oceanogr. Meth. 4, 346–357. doi: 10.4319/lom.2006.4.346
Heuer, V. B., Inagaki, F., Morono, Y., Kubo, Y., Spivack, A. J., Viehweger, B., et al. (2020). Temperature limits to deep subseafloor life in the Nankai trough subduction zone. Science 370, 1230–1234. doi: 10.1126/science.abd7934
Hoehler, T. M., Alperin, M. J., Albert, D. B., and Martens, C. S. (2001). Apparent minimum free energy requirements for methanogenic archaea and sulfate-reducing bacteria in an anoxic marine sediment. FEMS Microbiol. Ecol. 38, 33–41. doi: 10.1111/j.1574-6941.2001.tb00879.x
Holmes, D. E., Shrestha, P. M., Walker, D. J. F., Dang, Y., Nevin, K. P., Woodard, T. L., et al. (2017). Metatranscriptomic evidence for direct interspecies electron transfer between Geobacter and Methanothrix species in methanogenic rice paddy soils. Appl. Environ. Microbiol. 83, e00223–e00217. doi: 10.1128/AEM.00223-17
Hoshino, T., Doi, H., Uramoto, G.-I., Wörmer, L., Adhikari, R. R., Xiao, N., et al. (2020). Global diversity of microbial communities in marine sediment. Proc. Natl. Acad. Sci. U. S. A. 117, 27587–27597. doi: 10.1073/pnas.1919139117
House, C. H., Cragg, B. A., and Teske, A., the Leg 201 Scientific Party (2003). “Drilling contamination tests during ODP leg 201 using chemical and particulate tracers” in ODP Leg 201, eds. D’Hondt, S. L. Jørgensen, B. B. Miller, D. J. et. al. Proc. ODP, Init. Repts., 201 [Online]. Available at: http://www-odp.tamu.edu/publications/201_IR/chap_02/chap_02.htm
House, C. H., Orphan, V. J., Turk, K. A., Thomas, B., Pernthaler, A., Vrentas, J. M., et al. (2009). Extensive carbon isotopic heterogeneity among methane seep microbiota. Environ. Microbiol. 11, 2207–2215. doi: 10.1111/j.1462-2920.2009.01934.x
Inagaki, F., Hinrichs, K.-U., Kubo, Y., Bowles, M. W., Heuer, V. B., Hong, W. L., et al. (2015). Exploring deep microbial life in coal-bearing sediment down to ~2.5 km below the ocean floor. Science 349, 420–424. doi: 10.1126/science.aaa6882
Inagaki, F., Nunoura, T., Nakagawa, S., Teske, A., Lever, M., Lauer, A., et al. (2006). Biogeographical distribution and diversity of microbes in methane-bearing deep marine sediments on the Pacific Ocean margin. Proc. Natl. Acad. U. S. A. 103, 2815–2820. doi: 10.1073/pnas.0511033103
Kallmeyer, J., Pockalny, R., Adhikari, R. R., Smith, D. C., and D’Hondt, S. (2012). Global distribution of microbial abundance and biomass in subseafloor sediment. Proc. Natl. Acad. Sci. U. S. A. 109, 16213–16216. doi: 10.1073/pnas.1203849109
Kellermann, M. Y., Wegener, G., Elvert, M., Yoshinaga, M. Y., Lin, Y.-S., Holler, T., et al. (2012). Autotrophy as a predominant mode of carbon fixation in anaerobic methane-oxidizing microbial communities. Proc. Natl. Acad. Sci. U. S. A. 109, 19321–19326. doi: 10.1073/pnas.1208795109
Kelley, D. S., Karson, J. A., Früh-Green, G. L., Yoerger, D. R., Shank, T. M., Butterfield, D. A., et al. (2005). A serpentinite-hosted ecosystem: the lost City hydrothermal field. Science 307, 1428–1434. doi: 10.1126/science.1102556
Knittel, K., and Boetius, A. (2009). Anaerobic oxidation of methane: progress with an unknown process. Annu. Rev. Microbiol. 63, 311–334. doi: 10.1146/annurev.micro.61.080706.093130
Kormas, K. A., Meziti, A., Dählmann, A., de Lange, G. J., and Lykousis, V. (2005). Characterization of methanogenic and prokaryotic assemblages based on mcrA and 16S rRNA gene diversity in sediments of the Kazan mud volcano (Mediterranean Sea). Geobiology 6, 450–460. doi: 10.1111/j.1472-4669.2008.00172.x
Krzycki, J. A., Kenealy, W. R., DeNiro, M. J., and Zeikus, J. G. (1987). Stable carbon isotope fractionation by Methanosarcina barkeri during methanogenesis from acetate, methanol, or carbon dioxide-hydrogen. Appl. Environ. Microbiol. 53, 2597–2599. doi: 10.1128/aem.53.10.2597-2599.1987
Laso-Pérez, R., Wu, F., Crémière, A., Speth, D. R., Magyar, J. S., Zhao, K., et al. (2023). Evolutionary diversification of methanotrophic ANME-1 archaea and their expansive virome. Nature Microbiol. 8, 231–245. doi: 10.1038/s41564-022-01297-4
Lever, M. A. (2013). Functional gene surveys from ocean drilling expeditions – a review and perspective. FEMS Microbiol. Ecol. 84, 1–23. doi: 10.1111/1574-6941.12051
Lever, M. A., Alperin, M. A., Engelen, B., Inagaki, F., Nakagawa, S., Steinsbu, B., et al. (2006). Trends in basalt and sediment core contamination during IODP expedition 301. Geomicrobiol J. 23, 517–530. doi: 10.1080/01490450600897245
Lever, M. A., Heuer, V. B., Morono, Y., Masui, N., Schmidt, F., Alperin, M. J., et al. (2010). Acetogenesis in deep subseafloor sediments of the Juan de Fuca ridge flank: a synthesis of geochemical, thermodynamic, and gene-based evidence. Geomicrobiol J. 27, 183–211. doi: 10.1080/01490450903456681
Lever, M. A., Rogers, K., Lloyd, K. G., Overmann, J. O., Schink, B., Thauer, R. K., et al. (2015). Microbial life under extreme energy limitation: a synthesis of laboratory- and field-based investigations. FEMS Microbiol. Rev. 39, 688–728. doi: 10.1093/femsre/fuv020
Lever, M. A., and Teske, A. P. (2015). Diversity of methane-cycling archaea in hydrothermal sediment investigated by general and group-specific PCR primers. Appl. Environ. Microbiol. 81, 1426–1441. doi: 10.1128/AEM.03588-14
Lloyd, K. G., Alperin, M. J., and Teske, A. (2011). Environmental evidence for net methane production and oxidation in putative anaerobic Methanotrophic (ANME) archaea. Environ. Microbiol. 13, 2548–2564. doi: 10.1111/j.1462-2920.2011.02526.x
Ludwig, W., Strunk, O., Westram, R., Richter, L., Meier, H., Yadhukumar,, et al. (2004). ARB: a software environment for sequence data. Nucleic Acids Res. 32, 1363–1371. doi: 10.1093/nar/gkh293
Lynch-Stieglitz, J., Stocker, T. F., Broecker, W. S., and Fairbanks, R. G. (1995). The influence of air-sea exchange on the isotopic composition of oceanic carbon: observations and modeling. Global Biogeochem. Cycles 9, 653–665. doi: 10.1029/95GB02574
Marshall, I. P. G., Karst, S. M., Nielsen, P. H., and Jørgensen, B. B. (2018). Metagenomes from deep Baltic Sea sediments reveal how past and present environmental conditions determine microbial community composition. Mar. Genomics 37, 58–68. doi: 10.1016/j.margen.2017.08.004
Mauclaire, L., Zepp, K., Meister, P., and McKenzie, J. (2005). Direct in situ detection of cells in deep-sea sediment cores from the Peru margin (ODP leg 201, site 1229). Geobiology 2, 217–223. doi: 10.1111/j.1472-4677.2004.00035.x
Mayumi, D., Mochimaru, H., Tamaki, H., Yamamoto, K., Yoshioka, H., Suzuki, Y., et al. (2016). Methane production from coal by a single methanogen. Science 354, 222–225. doi: 10.1126/science.aaf8821
Meister, P., Prokopenko, M., Skilbeck, C. G., Watson, M., and McKenzie, J. A. (2005). “Data report: compilation of total organic and inorganic carbon data from Peru margin and eastern equatorial Pacific drill sites (ODP legs 112, 138, and 201)” in, eds. Jørgensen, B. B. D’Hondt, S. L. and Miller, D. J. Proc. ODP, Sci. Results, 201, 1–20 [Online]. Available at: http://www-odp.tamu.edu/publications/201_SR/VOLUME/CHAPTERS/105.PDF
Millero, F. J. (2000). The activity coefficients of non-electrolytes in seawater. Marine Chem. 70, 5–22. doi: 10.1016/S0304-4203(00)00011-6
Millero, F. J., and Schreiber, D. R. (1983). Use of the ion pairing model to estimate activity coefficients of the ionic components of natural waters. Am. J. Sci. 282, 1508–1540. doi: 10.2475/ajs.282.9.1508
Nauhaus, K., Albrecht, M., Elvert, M., Boetius, A., and Widdel, F. (2007). In vitro cell growth of marine archaeal-bacterial consortia during anaerobic oxidation of methane with sulfate. Environ. Microbiol. 9, 187–196. doi: 10.1111/j.1462-2920.2006.01127.x
Parkes, R. J., Cragg, B. A., Bale, S. J., Getliff, J. M., Goodman, K., Rochelle, P. A., et al. (2005). Deep sub-seafloor prokaryotes stimulated at interfaces over geological time. Nature 436, 390–394. doi: 10.1038/nature03796
Parkes, R. J., Cragg, B., Roussel, E., Webster, G., Weightman, A., and Sass, H. (2014). A review of prokaryotic populations and processes in sub-seafloor sediments, including biosphere: geosphere interactions. Mar. Geol. 352, 409–425. doi: 10.1016/j.margeo.2014.02.009
Penning, H., Claus, P., Casper, P., and Ralf Conrad, R. (2005). Carbon isotope fractionation during acetoclastic methanogenesis by Methanosaeta concilii in culture and a lake sediment. Appl. Environ. Microbiol. 72, 5648–5652. doi: 10.1128/AEM.00727-06
Rotaru, A.-E., Posth, N. R., Löscher, C. R., Miracle, M. R., Vicente, E., Cox, R. P., et al. (2019). Interspecies interactions mediated by conductive minerals in the sediments of the iron-rich meromictic Lake La Cruz, Spain. Limnetica 38, 21–40. doi: 10.23818/limn.38.10
Rotaru, A.-E., Shrestha, P. M., Liu, F., Shrestha, M., Shrestha, D., Embree, M., et al. (2014). A new model for electron flow during anaerobic digestion: direct interspecies electron transfer to Methanosaeta for the reduction of carbon dioxide to methane. Energy Environ. Sci. 7, 408–415. doi: 10.1039/C3EE42189A
Roussel, E. G., Bonavita, M. A. C., Querellou, J., Cragg, B. A., and Webster, G. (2008). Extending the sub-sea-floor biosphere. Science 320:1046. doi: 10.1126/science.1154545
Schink, B., Montag, D., Keller, A., and Müller, N. (2017). Hydrogen or formate: alternative key players in methanogenic degradation. Environ. Microbiol. Repts. 9, 189–202. doi: 10.1111/1758-2229.12524
Schippers, A., Kock, D., Höft, C., Köweker, G., and Siegert, M. (2012). Quantification of microbial communities in subsurface marine sediments of the Black Sea and off Namibia. Front. Microbiol. 3:16. doi: 10.3389/fmicb.2012.00016
Schippers, A., Neretin, L., Kallmeyer, J., Ferdelman, T., Cragg, B. A., Parkes, J. R., et al. (2005). Prokaryotic cells of the deep sub-seafloor biosphere identified as living bacteria. Nature 433, 861–864. doi: 10.1038/nature03302
Shipboard Scientific Party (1988). “Introduction, objectives, and principal results, leg 112, Peru continental margin” in E. Suess, R. von Huene, et al. Proc. ODP, Init. Repts., 112. (College Station, TX: Ocean Drilling Program), 5–23. doi: 10.2973/odp.proc.ir.112.102.1988
Smith, K. S., and Ingram-Smith, C. (2007). Methanosaeta, the forgotten methanogen? Trends Microbiol. 15, 150–155. doi: 10.1016/j.tim.2007.02.002
Sørensen, K. B., and Teske, A. (2006). Stratified communities of active Archaea in deep marine subsurface sediments. Appl. Environ. Microbiol. 72, 4596–4603. doi: 10.1128/AEM.00562-06
Sparling, R., and Daniels, L. (1986). Source of carbon and hydrogen in methane produced from formate by Methanococcus thermolithotrophicus. J. Bacteriol. 168, 1402–1407. doi: 10.1128/jb.168.3.1402-1407.1986
Spivack, A. J., McNeil, C., Holm, N. G., and Hinrichs, K.-U. (2005). “Determination of in situ methane based on analysis of void gas” in ODP Leg 201, eds. B. B. Jørgensen, S. L. D’Hondt, and D. J. Miller, Proc. ODP, Sci. Results, 201 [Online]. Available at: http://www-odp.tamu.edu/publications/201_SR/119/119.htm
Springer, E., Sachs, M. S., Woese, C. R., and Boone, D. R. (1995). Partial gene sequences for the a subunit of methyl-coenzyme M reductase (mcrI) as a phylogenetic tool for the family Methanosarcinaceae. Int. J. Syst. Bacteriol. 45, 554–559. doi: 10.1099/00207713-45-3-554
Stahl, D. A., and Amann, R. (1991). “Development and application of nucleic acid probes” in Nucleic acid techniques in bacterial systematics. eds. E. Stackebrandt and M. Goodfellow, vol. 8 (London: John Wiley & Sons), 207–248.
Stumm, W., and Morgan, J. J. (1996). Aquatic chemistry, chemical equilibria and rates in natural waters, 3rd ed. New York: John Wiley & Sons, Inc.
Suess, E. (1981). Phosphate regeneration from sediment of the Peru continental margin by dissolution of fish debris. Geochim. Cosmochim. Acta 45, 577–588. doi: 10.1016/0016-7037(81)90191-5
Sun, R., and Duan, Z. (2007). An accurate model to predict the thermodynamic stability of methane hydrate and methane solubility in marine environments. Chem. Geol. 244, 248–262. doi: 10.1016/j.chemgeo.2007.06.021
Thauer, R. K. (1998). Biochemistry of methanogenesis: a tribute to Marjory Stephenson. Microbiology 144, 2377–2406. doi: 10.1099/00221287-144-9-2377
Torti, A., Jørgensen, B. B., and Lever, M. A. (2018). Preservation of microbial DNA in marine sediments: insights from extracellular DNA pools. Environ. Microbiol. 20, 4526–4542. doi: 10.1111/1462-2920.14401
Wagner, M., Loy, A., Klein, M., Lee, N., Ramsing, N. B., Stahl, D. A., et al. (2005). Functional marker genes for identification of sulfate-reducing prokaryotes. Meth. Enzymol. 397, 469–489. doi: 10.1016/S0076-6879(05)97029-8
Wang, Y., Wegener, G., Ruff, S. E., and Wang, F. (2021). Methyl/alkyl-coenzyme M reductase-based anaerobic alkane oxidation in archaea. Environ. Microbiol. 23, 530–541. doi: 10.1111/1462-2920.15057
Webster, G., Parkes, R. J., Cragg, B. A., Newberry, C. J., Weightman, A. J., and Fry, J. C. (2006). Prokaryotic community composition and biogeochemical processes in deep subseafloor sediments from the Peru margin. FEMS Microbiol. Ecol. 58, 65–85. doi: 10.1111/j.1574-6941.2006.00147.x
Wegener, G., Gropp, J., Taubner, H., Halevy, I., and Elvert, M. (2021). Sulfate-dependent reversibility of intracellular reactions explains the opposing effects in the anaerobic oxidation of methane. Sci. Adv. 7:eabe4939. doi: 10.1126/sciadv.abe4939
Wegener, G., Niemann, H., Elvert, M., Hinrichs, K.-U., and Boetius, A. (2008). Assimilation of methane and inorganic carbon by microbial communities mediating the anaerobic oxidation of methane. Environ. Microbiol. 10, 2287–2298. doi: 10.1111/j.1462-2920.2008.01653.x
Whiticar, M. J. (1999). Carbon and hydrogen isotope systematics of bacterial formation and oxidation of methane. Chem. Geol. 161, 291–314. doi: 10.1016/S0009-2541(99)00092-3
Whiticar, M. J., Faber, E., and Schoell, M. (1986). Biogenic methane formation in marine and freshwater environments: CO2 reduction vs. acetate fermentation – isotope evidence. Geochim. Cosmochim. Acta 50, 693–709. doi: 10.1016/0016-7037(86)90346-7
Whitman, W. B., Bowen, T. L., and Boone, D. R. (2014). “The methanogenic Bacteria” in The prokaryotes. eds. E. Rosenberg, E. F. DeLong, S. Lory, E. Stackebrandt, and F. Thompson, vol. 3, (Heidelberg:Springer, Berlin) 124–163. doi: 10.1007/978-3-642-38954-2_407
Yang, P., Amy Tan, G.-Y., Aslam, M., Kim, J., and Lee, P.-H. (2019). Metatranscriptomic evidence for classical and RuBisCO-mediated CO2 reduction to methane facilitated by direct interspecies electron transfer in a methanogenic system. Sci. Rep. 9:4116. doi: 10.1038/s41598-019-40830-0
Yoshinaga, M. Y., Holler, T., Goldhammer, T., Wegener, G., Pohlman, J. W., Brunner, B., et al. (2014). Carbon isotope equilibration during sulphate-limited anaerobic oxidation of methane. Nat. Geosci. 7, 190–194. doi: 10.1038/ngeo2069
Keywords: deep biosphere, methanogenesis, anaerobic oxidation of methane, subseafloor sediment, ocean drilling, methane hydrate, carbon isotopes, mcrA
Citation: Lever MA, Alperin MJ, Hinrichs K-U and Teske A (2023) Zonation of the active methane-cycling community in deep subsurface sediments of the Peru trench. Front. Microbiol. 14:1192029. doi: 10.3389/fmicb.2023.1192029
Edited by:
Anirban Chakraborty, Idaho State University, United StatesReviewed by:
Xiyang Dong, Third Institute of Oceanography of the Ministry of Natural Resources, ChinaSabrina Beckmann, Oklahoma State University, United States
Copyright © 2023 Lever, Alperin, Hinrichs and Teske. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mark A. Lever, mark.lever@austin.utexas.edu
 Mark A. Lever
Mark A. Lever Marc J. Alperin
Marc J. Alperin Kai-Uwe Hinrichs
Kai-Uwe Hinrichs Andreas Teske
Andreas Teske