Abstract
Albidopyrone, a new α-pyrone-containing secondary metabolite, was produced by Streptomyces sp. NTK 227, a strain isolated from Atlantic Ocean sediment and found to be a member of the Streptomyces albidoflavus 16S rRNA gene clade. The structure of the compound was determined by MS and NMR spectroscopy, and found to have a moderate inhibitory activity against protein-tyrosin phosphatase B.
Similar content being viewed by others
Introduction
In our HPLC–diode array screening program to detect novel secondary metabolites from actinomycetes isolated from marine sediments,1, 2 extracts of culture filtrates of the strain NTK 227 were found to be of special interest. This organism, which was isolated from a marine sediment, produced a main metabolite in HPLC that runs at a retention time of 6.4 min in a standardized gradient elution profile (Figure 1). The characteristic UV–visible (Vis) spectrum of the metabolite differed from those of 867 reference compounds stored in our HPLC–UV–Vis database.3 The metabolite was characterized as a new member of the α-pyrone family by MS and NMR spectroscopy and was named albidopyrone (1); its structure is shown in Figure 2. Furthermore, metabolites in the culture filtrate extract of strain NTK 227 were identified by an HPLC–diode array analysis as ferulic acid (retention time 5.9 min) and fredericamycin A (retention time 11.4 min). In the mycelium extract, several members of the antimycin A complex and two representatives of yet non-characterized heptane macrolides were monitored. This study describes the taxonomy of the producing strain, and the fermentation, isolation and structural elucidation of albidopyrone, as well as on its biological activity.
Results
Taxonomy of the producing strain
Strain NTK 227 contained LL-diaminopimelic acid, hydrogenated menaquinones with nine isoprene units, produced a yellow white aerial spore mass and a brown diffusible pigment on oatmeal agar, and formed straight chains of smooth surfaced spores, properties typical of members classified in the genus Streptomyces.4 The organism formed a distinct phyletic line in the Streptomyces albidoflavus 16S rRNA gene clade,5 which was most closely related to the type strains of S. albidoflavus, S. canescens, S. coelicolor, S. felleus, S. limosus and S. odiferus sharing a 16S rRNA similarity with them of 99.9%, a value corresponding to eight nucleotide differences at 1429 locations; these values are consistent with the strain being seen as a new species.
Fermentation and isolation
When grown in a 10-l stirred tank fermentor, strain NTK 227 reached a maximal biomass of 8.5 mg dry weight/ml after incubation for 10 days. Production of 1 started at 72 h, reaching a maximal yield of 56 mg l−1 after an incubation period of 14 days.
Structure 1 was isolated from the mycelium by extraction with MeOH–Me2CO. The extracts were concentrated to an aqueous residue, combined with the culture filtrate and separated by Amberlite XAD-16 chromatography. After extraction with n-BuOH, the crude product was purified by preparative reversed-phase HPLC. Structure 1 was obtained as a white powder after concentration to dryness.
Structural elucidation
The mass spectrum derived from HPLC-ESI-MS chromatograms revealed the molecular mass for 1 [(M+H)+=260.2]. The exact molecular mass was determined by high-resolution ESI-FT-ICR-MS as 260.09161 Da [(M+H)+] (1), this corresponds to the molecular formula C14H13NO4 (1) [(M+H)+theor=260.09173; Δm=0.46 p.p.m.].
The 1H-NMR-spectrum of 1 showed two signals in the aromatic region, one signal at 10.2 p.p.m., one broad signal at 11.28 p.p.m. and two signals in the aliphatic region. The integration of the signals revealed that one of the two signals in the aromatic region corresponded to four protons. The other signal in the aromatic region, as well as the two signals at 10.2 p.p.m. and at 11.28 p.p.m., correspond each to one proton, and each of the two signals in the aliphatic region to three protons suggesting that two methyl groups are present in the compound (Table 1). Remarkably, the 13C-NMR spectrum showed 12 signals instead of the expected 14 signals (Table 1). Comparing the 13C-spectrum with the DEPT spectrum confirmed the assumption that two methyl groups are present in the compound. Furthermore, the DEPT spectrum showed three signals between 95 and 130 p.p.m.
The correlation of 1H-NMR signals to the corresponding C-atoms was carried out by means of interpretation of the Heteronuclear Multiple Quantum Coherence (HMQC) NMR experiment. In accordance with the DEPT spectrum, five signals were found. The signal in the aromatic region, which corresponds to four protons, showed correlations to only two carbons. This led to the conclusion that the two of them are chemically equivalent, suggesting symmetry for this part of the molecule, and thus explains the absence of two signals in the 13C-spectrum. The signals at 10.2 p.p.m. and at 11.28 p.p.m. in the 1H-spectrum could not be assigned to any C-atom, suggesting the presence of two heteroatom-bound protons (OH or NH). Interestingly, no correlation was seen in the 1H-1H-COSY experiment. Nevertheless, the structure could be fully elucidated using the HMBC spectrum. The chemical shifts in combination with the correlations from H-5 to C-3, C-4, C-6 and C-7; from H-8 to C-6, C-8 and C-10; from H-9 to C-7 and C-9; from H-11 to C-9 and C-12; from H-13 to C-12; and from H-14 to C-2, C-3 and C-4, shown in Figure 3, provided proof for the structure of 1, particularly for the position of the methyl groups, of the amide and hydroxy function, respectively.
The physico-chemical properties of albidopyrone (1) are summarized in Table 2.
Biological activity
Albidopyrone (1) did not show any growth inhibitory activity either against the Gram-negative bacteria (Escherichia coli and Pseudomonas fluorescens) and Gram-positive bacteria (Bacillus subtilis and S. lentus) or against the yeast (Candida glabrata). Structure 1 inhibited the PTP1B with an IC50 of 128 μg ml−1.
Discussion
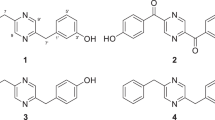
Herein we report on the fermentation, isolation, structural elucidation and the evaluation of the biological activity of albidopyrone (1), a metabolite that contains a 4-hydroxy-2-pyrone unit as its characteristic structural feature. α-Pyrones represent a class of six-membered lactones that are sub-structures of various natural products and which are highly abundant in animals, bacteria, insects and plants.6 They exhibit a wide range of biological activities, such as antifungal, cytotoxic, neurotoxic and phytotoxic properties.6, 7 The structural highly related benzo-α-pyrones 2, 3, 4 and 5 (Figure 4) have been synthesized, and their biological activities evaluated to some extent.6, 7, 8, 9, 10 The competitive inhibition of HIV-1 protease by 2, 4 and some other derivatives has been explored, revealing that the pyran-2-one group, the 4-hydroxyl group, and the substitution at position 3 were necessary for inhibitory activity.9 Moreover, 4-hydroxy-2-pyrones have become one of the most important classes of anti-HIV agents in recent years.6 These non-peptide compounds seem to be promising candidates for treatment of AIDS. In comparison with peptides, the small α-pyrones are interesting lead candidates for synthetic strategies because of their lack of chiral centers.6, 9 Recently, Lin et al.11 isolated two new 5-hydroxy-2-pyrone derivatives from a marine Aspergillus flavus strain, which might prove to be ligands for G-protein-coupled receptors.
Some structurally related benzo- and phenyl-α-pyrones have been isolated from plants, such as 6, 7 and 8; the structures of these compounds are shown in Figure 4. Pogopyrone B (6) was isolated from Pogostemon heynianus,12, 13 whereas 6-(2-hydroxyphenyl)-4-methoxy-2-pyrone (7) was isolated in a bioassay-guided purification of ether extracts from Alpinia officinarum,14 this metabolite exhibited a potent platelet-activating factor receptor-binding inhibitory activity. The aloenin aglycon (8) was isolated from the leaf exudate of Aloe nyeriensis var. kedongensis along with the O-glycosylated aloenin.15 The hitherto unknown albidopyrone (1) extends the structural diversity of this group of natural products. In particular, the amide function at C-10 has never been reported in related compounds neither in synthetic nor in natural products.
Albidopyrone (1) inhibited specifically the PTP1B, other biological activities have yet to be found. Protein tyrosine phosphatases and their counterpart protein tyrosine kinases leading to reversible protein tyrosine phosphorylation are of importance in the regulation of various cellular signal transduction systems. PTP1B is the major negative regulator of insulin signaling because it regulates the phosphorylation state of the insulin receptor or insulin receptor substrate.16 Type II diabetes and obesity are characterized by resistance to insulin because of attenuated or diminished signaling from the receptors. Therefore, pharmacological agents capable of inhibiting PTP1B can increase the action of insulin, and thus serve as a very attractive target for treatment of both type II diabetes and obesity and various inhibitors were identified.16, 17
Materials and methods
Producing organism and taxonomy
Strain NTK 227 was isolated from Atlantic Ocean sediment. The organism was examined for chemotaxonomic and morphological properties known to be of value in streptomycete systematics.18, 19 16S rRNA gene amplification and sequencing were carried out using an established method,20 and the resultant almost complete sequence compared with corresponding sequences of Streptomyces type strains using the neighbor-joining algorithm.21
Fermentation and isolation
Batch fermentations of strain NTK 227 were carried out in a 10-liter stirred tank fermentor (Biostat E; B Braun, Melsungen, Germany) in a complex medium that consisted of (per liter tap water) oatmeal (Holo Hafergold, Neuform, Germany) 20 g and trace element solution 5 ml, which was composed of (per liter deionized water) CaCl2 × 2H2O 3 g, iron(III) citrate 1 g, MnSO4 × 1H2O 200 mg, ZnCl2 100 mg, CuSO4 × 5H2O 25 mg, Na2B4O7 × 10H2O 20 mg, CoCl2 × 6H2O 4 mg and Na2MoO4 × 2H2O 10 mg; the pH was adjusted to 7.3 (5 M HCl) before sterilization. The fermentor was inoculated with 5% by volume of a shake flask culture grown in a seed medium at 27 °C in 500-ml Erlenmeyer flasks with a single baffle for 72 h on a rotary shaker at 120 r.p.m. The seed medium consisted of glucose 10 g, glycerol 10 g, oatmeal 5 g, soybean meal (Schoenenberger, Magstadt, Germany) 10 g, yeast extract (Ohly Kat, Deutsche Hefewerke, Hamburg, Germany) 5 g, Bacto casamino acids 5 g and CaCO3 1 g in 1 l tap water. The fermentation was carried out for 14 days at an aeration rate of 0.5 volume air/volume/min and with agitation at 250 r.p.m. The pH was kept constant at 7.0 during the whole fermentation period.
Hyphlo Super-cel (2%) was added to the fermentation broth, and the resultant preparation was separated by multiple sheet filtration into culture filtrate and mycelium. The mycelium was extracted three times with 1 l MeOH–Me2CO (1:1). The mycelium extracts were concentrated in vacuo to an aqueous residue (500 ml), combined with the culture filtrate (6 l) and applied to an Amberlite XAD-16 column (resin volume 800 ml). The resin was washed with each of 3.2 l H2O and H2O–MeOH (8:2); albidopyrone (1) was eluted with 2.4 l H2O–MeOH (2:8) and concentrated in vacuo to an aqueous residue. After defatting with 250 ml petroleum benzene (40–60 °C), the concentrate was extracted six times with n-BuOH (1 l in total), and the organic extracts combined and concentrated in vacuo to dryness (3.4 g). The crude product was dissolved in DMSO; 1 ml of this preparation was diluted with the same volume H2O directly before separation and subject to preparative HPLC (LaPrep P110; VWR, Darmstadt, Germany) using a C18 column (Nucleosil 100 C-18, 10 μm, 250 × 16 mm; Maisch, Ammerbuch, Germany) with 0.5% HCOOH–MeOH (a linear gradient from 30 to 50% MeOH over 15 min) at a flow rate of 24 ml min−1. The eluate was monitored at 240 and 360 nm. Structure 1-containing fractions were combined and concentrated in vacuo to dryness and a white powder (130 mg) was obtained.
HPLC–diode array detection analyses
The chromatographic system consisted of an HP 1090M liquid chromatograph equipped with a diode-array detector and an HP Kayak XM 600 ChemStation (Agilent, Waldbronn, Germany). Multiple wavelength monitoring was performed at 210, 230, 260, 280, 310, 360, 435 and 500 nm, and UV–Vis spectra measured from 200 to 600 nm. A 10-ml aliquot of the fermentation broth was centrifuged, and the supernatant adjusted to pH 4.0 and extracted with the same volume of EtOAc. After centrifugation, the organic layer was concentrated to dryness in vacuo and resuspended in 1 ml MeOH. Aliquots of the samples, in volumes of 10 μl, were injected into an HPLC column (125 × 4.6 mm) fitted with a guard-column (20 × 4.6 mm) filled with 5-μm Nucleosil-100 C-18 (Maisch). The samples were analyzed by linear gradient elution using 0.1% ortho-phosphoric acid as solvent A and MeCN as solvent B at a flow rate of 2 ml min−1. The gradient was from 0 to 100% for solvent B in 15 min with a 2-min hold at 100% for solvent B.
Structural elucidation
LC-MS experiments were performed on an Applied Biosystems QTrap 2000 (Applied Biosystems, Darmstadt, Germany) coupled to an Agilent 1100 HPLC system (Agilent). High-resolution ESI-FT-ICR mass spectra were recorded on an APEX II FTICR mass spectrometer (4.7 T; Bruker-Daltronics, Bremen, Germany) and NMR experiments on a DRX 500 NMR spectrometer (Bruker, Karlsruhe, Germany) equipped with a BBI probe head with z gradients. DMSO-d6 was used as a solvent for NMR experiments and chemical shifts are referenced to tetramethylsilane.
Biological activity
An agar plate diffusion assay was used to determine the antibacterial and antifungal properties of albidopyrone (1) using Escherichia coli K12 (DSM 498), Pseudomonas fluorescens (NCIMB 10586), Bacillus subtilis (DSM 347), Staphylococcus lentus (DSM 6672) and Candida glabrata (DSM 6425).
Analysis of the effect of 1 on human recombinant protein tyrosine phosphatase 1B (PTP1B) was carried out with final concentrations of the substance of 20 μg ml−1 in PTP1B assay buffer containing 100 mM Hepes buffer (pH 7.2), 2 mM EDTA, 2 mM dithiothreitol and 0.1% nonylphenylpolyethylene glycol (NP-40) (Cat. no. KI-131; Biomol, Hamburg, Germany), 5 ng bovine serum albumin and 3 ng (150 U (150 pmol min−1)) recombinant human PTP1B (Cat. no. SE332-0050; Biomol) in a volume of 45 μl per well. The reaction was started with 5 μl of the 1.5 mM PTP1B phosphopeptide substrate EGFR (988-998) (Cat. no. P323-0001; Biomol) dissolved in PTP1B assay buffer. After an incubation period of 15 min at 30 °C, the reaction was stopped by adding 100 μl of Biomol Green (Cat. no. AK111-9090; Biomol); the ortho-phosphate concentration was quantified after incubating for 20 min at room temperature. Optimal density was measured at 620 nm using the microtiter plate reader Infinite M200 (Tecan). As a positive control for inhibition of PTP1B, 200 μM of RK-682 (Cat. no. 557322-200UG; Calbiochem ) was added instead of the test substance.
References
Hohmann, C. et al. Caboxamycin, a new antibiotic of the benzoxazole family, produced by the deep-sea strain Streptomyces sp. NTK 937. J. Antibiot., in press.
Fiedler, H.-P. et al. Marine actinomycetes as a source of novel secondary metabolites. Antonie van Leeuwenhoek 87, 37–42 (2005).
Fiedler, H.-P. Biosynthetic capacities of actinomycetes. 1. Screening for novel secondary metabolites by HPLC and UV–visible absorbance libraries. Nat. Prod. Lett. 2, 119–128 (1993).
Manfio, G. P., Zakrezewska-Czerwinska, J., Atalan, E. & Goodfellow, M. Towards minimal standards for the description of Streptomyces species. Bioteckhnologia 7–8, 242–253 (1995).
Lanoot, B. et al. Grouping of streptomycetes using 16S-ITS RFLP fingerprinting. Res. Microbiol. 156, 755–762 (2005).
McGlacken, G. & Fairlamb, I. J. S. 2-Pyrone natural products and mimetics: isolation, characterisation and biological activity. Nat. Prod. Rep. 22, 369–385 (2005).
Katritzky, A. R., Wang, Z., Wang, M., Hall, C. D. & Suzuki, K. Facile syntheses of 2,2-dimethyl-6-(2-oxoalkyl)-1,3-dioxin-4-ones and the corresponding 6-substituted 4-hydroxy-2-pyrones. J. Org. Chem. 70, 4854–4856 (2005).
Morris, J., Luke, G. P. & Wishka, D. G. Reaction of phosgeniminium salts with enolates derived from Lewis acid complexes of 2′-hydroxypropiophenones and related β-diketones. J. Org. Chem. 61, 3218–3220 (1996).
Tummino, P. J., Ferguson, D., Huper, L. & Hupe, D. Competitive inhibition of HIV-1 protease by 4-hydroxy-benzopyran-2-ones and by 4-hydroxy-6-phenylpyran-2-ones. Biochem. Biophys. Res. Commun. 200, 1658–1664 (1994).
Liebeskind, L. S. & Wang, J. A synthesis of substituted 2-pyrones by carbonylative cross-coupling-thermolysis of 4-halocyclobutenones with alkenyl-, aryl-, and heteroarylstannanes. Tetrahedron 49, 5461–5470 (1993).
Lin, A. et al. Two new 5-hydroxy-2-pyrone derivatives isolated from a marine-derived fungus Aspergillus flavus. J. Antibiot. 61, 245–249 (2008).
Thailambal, V. G. & Pattabhi, V. Structure of 3-acetyl-4-hydroxy-6-phenyl-2-pyrone, C13H10O4 . Acta Crystallogr. C 41, 802–804 (1985).
Purushothaman, K. K., Sarada, A. & Connolly, J. D. Structures of pogopyrones A and B. Indian J. Chem., Sect B, Org. Chem. Med. Chem. 23B, 611–614 (1984).
Fan, G., Kang, Y.-H., Han, Y. N. & Han, B. H. Platelet-activating factor (PAF) receptor binding antagonists from Alpinia officinarum. Bioorg. Med. Chem. Lett. 17, 6720–6722 (2007).
Conner, J. M., Gray, A. I., Reynolds, T. & Waterman, P. G. Anthraquinone, anthrone and phenylpyrone components of Aloe nyeriensis var. kedongensis leaf exudate. Phytochemistry 26, 2995–2997 (1987).
Zhang, S. & Zhang, Z.-Y. PTP1B as a drug target: recent developments in PTP1B inhibitor discovery. Drug Discov. Today 12, 373–381 (2007).
Taylor, S. D. Inhibitors of protein tyrosine phosphatase 1B (PTP1B). Curr. Top. Med. Chem. 3, 759–782 (2003).
Williams, S. T., Goodfellow, M. & Alderson, G. Genus Streptomyces Waksman and Henrici 1943, 339AL. in: Williams S.T., et al. Bergey's Manual of Systematic Bacteriology, Vol. 4. (eds Williams, S. T. et al.) (Williams & Wilkins, Baltimore, 1989).
Xu, C. et al. Neutrotolerant acidophilic Streptomyces species isolated from acidic soils in China: Streptomyces guanduensis sp.nov., Streptomyces paucisporus sp. nov., Streptomyces rubidus sp. nov. and Streptomyces yanglinensis sp. nov. Int. J. Syst. Evol. Microbiol. 56, 1109–1115 (2006).
Kim, S. B., Falconer, C., Williams, S. T. & Goodfellow, M. Streptomyces thermocarboxydovorans sp. nov. and Streptomyces thermocarboxydus sp. nov., two moderately thermophilic carboxydotrophic species from soil. J. Syst. Bacteriol. 48, 59–88 (1998).
Saitou, N. & Nei, M. The neighbour-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425 (1987).
Acknowledgements
This work was supported by Boehringer-Ingelheim Pharma GmbH (Biberach, Germany), the European Commission (project ACTINOGEN, 6th framework, Grant LSHM-CT-2004-005224). We thank Mr G Grewe, Universität Tübingen, for technical assistance in the fermentations, and Agilent Technologies (Waldbronn, Germany) for HPLC software support.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Art. no. 49 in ‘Biosynthetic Capacities of Actinomycetes’. Art. no. 48: see ref. 1.
Rights and permissions
About this article
Cite this article
Hohmann, C., Schneider, K., Bruntner, C. et al. Albidopyrone, a new α-pyrone-containing metabolite from marine-derived Streptomyces sp. NTK 227. J Antibiot 62, 75–79 (2009). https://doi.org/10.1038/ja.2008.15
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ja.2008.15
Keywords
This article is cited by
-
Multicomponent synthesis of a novel bicyclic pyran-2-one system
Monatshefte für Chemie - Chemical Monthly (2022)
-
Bioprospection of marine actinomycetes: recent advances, challenges and future perspectives
Acta Oceanologica Sinica (2019)
-
Natural products possessing protein tyrosine phosphatase 1B (PTP1B) inhibitory activity found in the last decades
Acta Pharmacologica Sinica (2012)