- 1Food Security and Safety Focus Area, Faculty of Natural and Agricultural Science, North-West University, Mafikeng, South Africa
- 2Division of Agrometeorology, Agricultural Research Council, Natural Resources and Engineering, Pretoria, South Africa
With the rise in the world population, environmental hazards caused by chemical fertilizers, and a decrease in food supply due to global climate change, food security has become very pertinent. In addition, considerable parts of agriculture lands have been lost to urbanization. It has therefore been projected that at the present rate of population increase coupled with the other mentioned factors, available food will not be enough to feed the world. Hence, drastic approach is needed to improve agriculture output as well as human sustainability. Application of environmentally sustainable approach, such as the use of beneficial microbes, and improved breeding of underutilized legumes are one of the proposed sustainable ways of achieving food security. Microbiome-assisted breeding in underutilized legumes is an untapped area with great capabilities to improve food security. Furthermore, revolution in genomics adaptation to crop improvement has changed the approach from conventional breeding to more advanced genomic-assisted breeding on the host plant and its microbiome. The use of rhizobacteria is very important to improving crop yield, especially rhizobacteria from legumes like Bambara groundnut (BGN). BGN is an important legume in sub-Saharan Africa with high ability to tolerate drought and thrive well in marginalized soils. BGN and its interaction with various rhizobacteria in the soil could play a vital role in crop production and protection. This review focus on the importance of genomics application to BGN and its microbiome with the view of setting a potential blueprint for improved BGN breeding through integration of beneficial bacteria.
Introduction
Today, there is an incisive reduction in crop production and diversity due to the inability to domesticate wild species, climate change impact, and urbanization (because of increasing population). For example, rice, wheat, and maize are the leading food crops in the world out of the thousands of the cultivable crop species available (Kumar et al., 2018). Furthermore, given the increasing world population, the production of sustainable food supplies will be a critical challenge in the 21st century. The world population is projected to cross 9 billion by 2050, indicating that food supplies must be doubled to meet the requirement of the expanding population (Laplaze et al., 2018). Beside increasing the quantity of food, improving quality is also critical to maintaining nutritive values. In addition to the mentioned problems, the use of chemical fertilizers in crop production has been a major cause of concern in their roles in increasing availability of chemical hazards in the environment. Hence, the need and advocation of an environmentally sustainable alternative to improving agricultural productivity has increased significantly.
Therefore, need for biodiversity in food crops and the incorporation of other lesser-known crops which are mostly referred to as underutilized/orphan crops, into the major food system is important now than ever. Various conventional and molecular breeding approaches have been employed to increase food production but there is still a very long way to go in meeting food demand. Molecular breeding methods have implemented the crop genome sequences as a key factor for understanding the processes (both physiological and biochemical) controlling plant traits and plant’s mode of responses to biotic and abiotic stresses. The rapid evolution of genome sequencing technologies has resulted in the generation of large data of plant genomes. This has created an opportunity for the application of this technology to crop improvement (Saima et al., 2017; Yuan et al., 2017) through various technologies. In the vastly improving area of life science technologies, new areas have merged for elucidating gene functions and metabolic pathways; “omics” technologies coupled with improved bioinformatics tools and databases. As our understanding of the key processes increases, it must be translated to researches in plant development and improved crop yield. Various next-generation-based technology has been developed to the generated large data sets. These protocols have been applied in genome-wide association studies (GWAS), quantitative linkage locus (QTLs) analysis, linkage mapping, genome selection, population genetics, marker-assisted breeding, genome editing, and SNP detections (Singh et al., 2011, 2017). All these protocols have been rightly optimized and modified for several model crops. A vast number of underutilized legumes have been proposed to help achieve food security especially in sub-Saharan Africa.
Legumes which are the most important food crops behind cereals belong to the family Leguminosae. They are important sources of proteins and minerals which makes them essential to poor people in underdeveloped communities most especially in Africa and Asia where the majority cannot afford meat and fish. Utilization of legumes in combatting malnutrition and food insecurity (Ojiewo et al., 2015; Mubaiwa et al., 2018; Olanrewaju et al., 2022) has been one of the focus of research in most developing countries. This focus outlines the visible potentials that need exploiting in these crops. Areas where they are majorly grown, include Nigeria, Senegal, Togo, Indonesia, Cameroun, India, and Cote d’Ivoire (Borget, 1992). They are classified as pulses (Bambara groundnut), oilseeds (Soybean and Groundnut), forage legumes (Winged bean), tuberous root (Yam bean), and food crops (Cowpea; Foyer et al., 2016; Mayes et al., 2019). Some are well known and extensively incorporated into the global food system while some are still relatively unknown and underutilized. Underutilization of these legumes can be because of a lack of knowledge/information on their uses and values. Among the underutilized legumes are African yam bean, Pigeon pea, and Bambara groundnut (BGN). Furthermore, a subset of these underutilized legumes perform better in marginal soils and under less favorable environmental conditions than their major crop counterparts (Olanrewaju et al., 2022). Hence, developing these subsets further for future agriculture makes a suitable and complementary approach to the continued use of major crops. This is particularly important given the expected negative impact of climate change on current major crop production systems and the gap between the current rate of genetic improvement of most major crops and the higher rates of food production required to be able to feed the predicted 9 billion in 2050 (Laplaze et al., 2018).
One of such underutilized crops Bambara groundnut [Vigna subterranea (L.) Verdc.], an indigenous African legume that thrives in marginalized soils where other crops do not thrive well (Halimi et al., 2019; Mayes et al., 2019; Olanrewaju et al., 2022). It is classified under the family Fabaceae, sub-family Faboidea, and the genus Vigna (Olanrewaju et al., 2022). There are two varieties known as the wild variety (Vigna subterranea var. spontanea) and the cultivated variety (Vigna subterranea var. subterranea; Yao et al., 2015). This legume seed crop was reported to originate from West Africa from the Bambara district near Timbuktu (Jideani and Diedericks, 2014) but is now widely grown in Africa, Malaysia, South, and Central America, some parts of Northern Australia, Sri Lanka, and Indonesia (Azam-Ali et al., 2001). Different places have their indigenous names for BGN for example in Malawi (Madagascar groundnut, Baffin pea, Voandzou, Indhlubu, underground bean, Nzama), Nigeria (Epa-Roro), South Africa (Jugo beans), and Zimbabwe (Nyimo beans; Emelike and Barber, 2018; Majola et al., 2021; Olanrewaju et al., 2022).
Its ability to fix nitrogen improves soil fertility and makes it useful in crop rotation and the possibility to be grown without the use of expensive chemicals and fertilizers (Babalola et al., 2017). Bearing in mind that chemicals and fertilizers are usually difficult to obtain in isolated areas, therefore, nitrogen fixation by BGN adds to its advantages to farmers especially in developing nations where it is cultivated. This crop has shown large diversity in its genetic resources for improvement and its richness in nutrient and mineral composition have been well-documented (Adebola et al., 2017; Atoyebi et al., 2017; Halimi et al., 2019; Valombola et al., 2019; Khan et al., 2020; Nyau et al., 2020; Adegboyega et al., 2021; Hlanga et al., 2021; Olanrewaju et al., 2021b).
Furthermore, BGN reportedly ticks the box for achieving food security crop (Massawe et al., 2016; Halimi et al., 2019; Olanrewaju et al., 2022). It is available, accessible, and affordable, and it is a source of income for farmers in sub-Sahara Africa due to its ability to tolerate drought and fix atmospheric nitrogen (Paliwal et al., 2020; Majola et al., 2021; Olanrewaju et al., 2021c). Furthermore, it cross-nodulates with nitrogen-fixing bacteria from other leguminous plants like cowpea (Laurette et al., 2015; Hassen et al., 2022) which further enhances its nitrogen-fixing ability. In sub-Saharan Africa, it is the third most eaten legume after groundnut (Arachis hypogea) and cowpea (Vigna unguiculata; Omoikhoje, 2008; Olukolu et al., 2012; Chai et al., 2017). It is nutritionally comparable to other legumes (Figure 1), such as soybeans, in the essential amino acids of lysine, methionine, and cysteine (Bamishaiye et al., 2011; Halimi et al., 2019). Compared to pigeon pea, lentils, and cowpea, BGN seed has a higher gross energy value (Hillocks et al., 2012). The protein of BGN seed competes favorably with other legumes, such as groundnut, cowpea, and pigeon pea, and it is found to be superior to the protein of other legumes (Halimi et al., 2019; Olanrewaju et al., 2022).
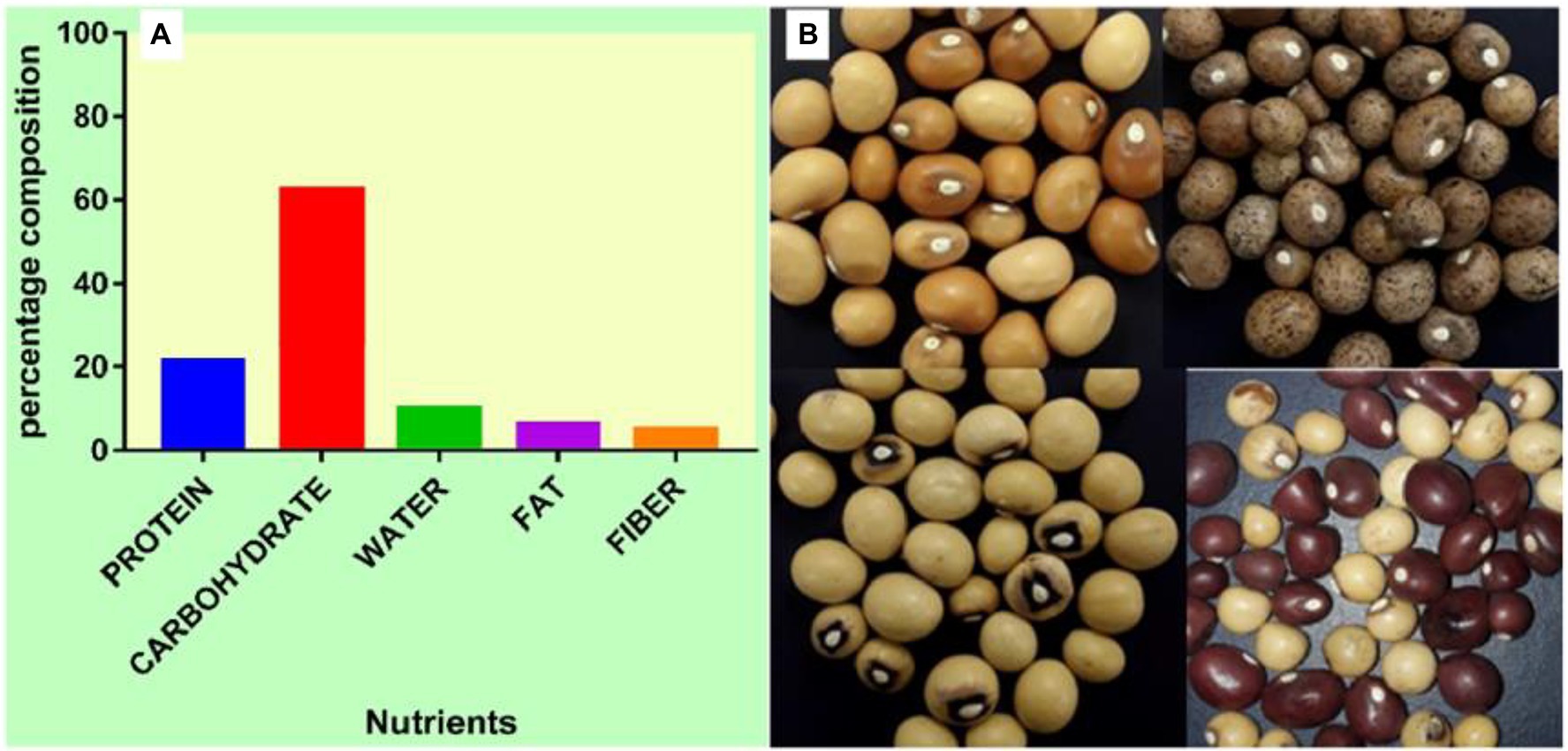
Figure 1. (A) Percentage nutrient composition of BGN. (B) Seeds of BGN showing variations in color, shape, and eye pattern.
It can be eaten boiled or milled into flour before cooking. In Senegal, different concoctions made from the leaves, roots, and leaf sap have been used to treat infected wounds and abscesses, as an aphrodisiac, and to treat epilepsy, respectively, (Brink et al., 2006). The plant has been used by the Ibo tribe in Nigeria to treat venereal disease, while the seeds that were pounded and mixed with water were used to treat cataracts. Its leaves were used as fodder to feed animals (Brink et al., 2006).
Crop diversification through the inclusion of underutilized legumes into major crop system will enhance food availability, however, activities in soil have been shown to have a great impact in crop production and response to environmental stress. Soil contains myriads of microorganisms which function in promoting plant growth through direct or indirect activities. Out of these microorganisms, bacteria are the most documented, both beneficial and pathogenic. The beneficial bacteria, referred to as plant growth-promoting rhizobacteria/bacteria (PGPR/B; Olanrewaju et al., 2017), have been a breakthrough in food production. The bacterial-mediated activities in the rhizosphere include those beneficial to humans, such as the key role they play in the biogeochemical cycles of the main elements (carbon, nitrogen, and sulfur) and of trace elements (Iron, Nickel, and Mercury). These activities increase the involvement of bacteria in energy exchanges within the soil (Haichar et al., 2014). Bacteria also synthesize vitamins, auxins, enzymes, and other growth factors for plant growth promotion and disease suppression (Olanrewaju et al., 2017). Bacterial communities in the rhizosphere are influenced by the soil characteristics, type of host plant, and host developmental stages (Chaparro et al., 2014; Anzuay et al., 2021). BGN and other underutilized legumes are critical for achieving food security, especially in sub-Saharan Africa.
No established varieties of BGN which means that the crop is cultivated from local landraces instead of having different varieties. The ability of breeding programs to harness the advantages of the new life science technologies which apply various molecular markers in gaining a better understanding of BGN genetics is a great step in the right direction. Due to the importance of this crop, these technologies can improve farmer’s income through improved crop yield as well as improving both global and local food security.
With the issue of underdevelopment and climate changes, there is no better solution to combat the looming food scarcity than to look within. Therefore, the improvement of indigenous crops, such as BGN, will proffer a lasting solution in a continent like Africa. Furthermore, not much work has been done on the bacterial community from the rhizosphere of BGN and its interactions with the plant itself. Hence, this review aims to outline the importance of beneficial microbes in improving BGN production and the outline the use of omics techniques as an effective means to characterize BGN-beneficial microbe interaction mechanisms. This will provide a theoretical basis for improving BGN breeding through the combined application of omics and plant–microbe interactions to achieve food security. The findings should prove useful in exploring mechanisms of the relationship between BGN and associated microbial species.
History of Bambara groundnut, Agronomy, and Morphology
The origin of BGN can be traced to Africa (Hillocks et al., 2012; Jideani and Jideani, 2021b; Soumare et al., 2021). Its history dates back to ancient Mali near Timbuctoo, from where the English name Bambara was derived from the Bambara tribe, even though they do not lay claim to the plant (Masindeni, 2006). Its center of origin can be traced to North Central and North Eastern Nigeria all the way to Northern Cameroon and the Central African Republic (Olukolu et al., 2012). Beyond Africa, it is seen to grow in other tropical nations like Greece, the Middle East, Malaysia, Indonesia, and, most especially, Brazil and Tropical America, where it is supposed that slaves must have helped to transport it to these nations (Brink et al., 2006).
It is an indigenous African crop that is common to many African countries, from Sudan in the North to South Africa in the South; from Kenya in the East to Nigeria in the West and even to Madagascar (Bamishaiye et al., 2011). It is one of the many underutilized and under-researched indigenous grain legumes. Female subsistence farmers in sub-Saharan Africa are the major growers of BGN (Mkandawire, 2007; Majola et al., 2021).
The annual production of BGN was estimated at 330,000 t in 1982 with about 50% coming from West Africa (William et al., 2016). In most of the semi-arid lands, yields from the farm pods vary between 650 and 850 kg ha−1 (Sangare, 2012). Since the crop is being produced at the subsistence level and not so much on a large scale, worldwide production figures have been difficult to collate but Zambia is still the most extensive producer (Opoku, 2010) while Nigeria, Burkina Faso, Niger, Mali, Ghana, Cote D’Ivoire and Chad are the major producers with only Burkina Faso, Chad, Mali, Niger and Senegal as the main exporting countries (Brink et al., 2006).
According to a report by Bamishaiye et al. (2011), BGN is a member of the family Fabaceae. It is a small plant-like groundnut which grows to a height of between 0.30–0.35 m. It is an intermediate plant with branched stems forming a bunch just above the ground. The plant grows as a small herb with compound leaves of three leaflets which are trifoliate and alternate from erect petioles. The peduncles of the leaves bear one or two flowers and are auxiliary branches from the stems. The stem begins to branch out very early after planting (Sangare, 2012). After fertilization, the flowers of BGN are pale yellow in color and they hang on the branching stems; these stems then grow downwards into the soil, taking the developing seed with it. The seeds form pods encasing seeds just below the ground in a similar fashion to peanuts. BGN pods are of different shapes, sizes, and colors, such as round, wrinkled, smooth, over 1.27 cm long, and white, cream, dark-brown, red, or black, and may be speckled or patterned with a combination of these colors, respectively (Figure 1). The roots with numerous nitrogen-fixing nodules grow from the short internodes of the stem to form a thick taproot with lateral roots developing as outgrowths toward the tip (Tweneboah, 2000). It is referred to as an autogamous plant (Baudoin and Mergeai, 2001). The structure of this plant shows that it helps to conserve space and more seeds can be planted on a small expanse of land without fear of low harvest. Its autogamy helps it to be available all through the year since it can be cultivated without the need for external pollination before flowering and seeding. Its availability throughout the year represents one of the core values of food security (Onwubiko et al., 2011).
Importance of Bambara Groundnut Cultivation
BGN is cultivated for various reasons. It is known to have both agronomic and nutritional advantages.
Agronomic Advantages of Bambara Groundnut
BGN is quite important in agriculture and a drought-tolerant crop (Masindeni, 2006; Babalola et al., 2017; Tan et al., 2020; Olanrewaju et al., 2021c; Soumare et al., 2021). It can perform well and have good crop yield on marginal soils and soils that have undergone water stress compared to other legumes (Brink et al., 2006). It grows well even in poor and infertile soils (Opoku, 2010; Olanrewaju et al., 2022). Because it tolerates poor soil, farmers with poor resources, especially with respect to purchasing fertilizer to increase yield, are encouraged to farm more with BGN. As a legume, BGN roots form a symbiotic association with root nodules of bacteria. This helps to increase the nitrogen content of the soil in the sense that the bacteria assimilate atmospheric nitrogen, trap it, and make it available to the plant in the soil (Babalola et al., 2017). This process in turn helps to increase soil fertility, which leads to increased crop yield (Masindeni, 2006). There are claims that it can reduce pests in the field (Ajayi and Lale, 2001; Udeh et al., 2020). It is however affected by storage pests which are the main cause of yield loss in most legume crops. These pests include cowpea weevil, bruchids, groundnut jassid, and brown leaf beetles (Majola et al., 2021).
Nutritional Advantages of Bambara Groundnut
BGN is a complete food having different composition of carbohydrate, protein, and fat enough to serve as a balanced diet (Ijarotimi and Esho, 2009; Halimi et al., 2019; Tan et al., 2020; Majola et al., 2021; Olanrewaju et al., 2022). Further processing by fermentation also improves its nutritive and mineral components (Murevanhema and Jideani, 2013). Compared to pigeon pea, lentils, and cowpea, BGN’s seed has a higher gross energy value (Bamishaiye et al., 2011). The protein content is found to be of a higher quality (16–25%) compared to other legumes (Adegboyega et al., 2021; Hlanga et al., 2021). Its carbohydrate and fat composition are 65 and 6.5%, respectively, (Mazahib et al., 2013). Its fat content is higher than that of cowpea (1.0 ± 1.6%) and pigeon pea (1.2 ± 1.5%) but lower than that of groundnut (peanut; 45.3 ± 47.7%) with an estimate of between 5% (Sangare, 2012) and 6.3% (Omoikhoje, 2008). The composition of its protein is superior in essential amino acids and includes phenylalanine, lysine, valine, methionine, leucine, threonine, and isoleucine. Its fatty acid composition is also high in palmitic, linolenic, and linoleic acids (Halimi et al., 2019). The protein in BGN is rich in lysine and methionine comprising 6.6 and 1.3% of the total protein, respectively. It is also a rich source of iron, potassium, calcium, and fiber (Omoikhoje, 2008; Hillocks et al., 2012).
Bambara Groundnut as Food
BGN is eaten when not matured by boiling it with salt and pepper. In many West African countries, it is consumed as a snack. It can also be made into flour when it is dry and mature because the seeds are hard (Tweneboah, 2000). The flour is used to prepare soup in East Africa with or without condiments while the flour has also been used to make bread in Zambia (Cook, 2017). The seeds can also be roasted, after which they are boiled, crushed, and eaten as a snack. Furthermore, the ground seeds can be used to make “akara” and “moinmoin” or the popular “okpa” in Nigeria (Emelike and Barber, 2018).
GIHOC cannery in Nsawam, Ghana, canned BGN seeds in gravy. Over 40, 000 cans of various sizes were prepared and made available throughout the year (Baudoin and Mergeai, 2001) and was comparable to Heinz baked beans even though its production declined due to competition with high yielding groundnut varieties and pest-resistant cowpea.
Vegetable milk extracted from BGN has been found to compete favorably with vegetable milk from soyabean, cowpea, and pigeon pea (Murevanhema and Jideani, 2013). The milk when properly processed has also been used as a weaning complementary food for children (Majola et al., 2021). The seeds were used as feed for poultry and piggery while its haulm and leaves which are rich in phosphorus and protein are used as fodder for cattle (Pui et al., 2021). BGN mixed with other leaf proteins has been used as an aquaculture feed with distinct growth in the fish (Adeparusi and Agbede, 2005).
Medicinal Importance of Bambara Groundnut
Different preparations from BGN have been shown to have medicinal properties (Murevanhema and Jideani, 2013). Preparations from the leaves have been applied to infected wounds and abscess; extracts from the leaves have been applied to the eyes to cure epilepsy (Khan et al., 2021) while leaf extracts pounded with that of Lanfana trifolia L. have been used as an insecticide to wash livestock (Mkandawire, 2007). Venereal disease is treated by the Igbo tribe in Nigeria using the plant (Oluwole et al., 2021). Grounded Bambara seeds when mixed with water have been used to treat cataracts in Senegal and the root has been used as an aphrodisiac (Brink et al., 2006). Water from boiled maize and BGN when drunk is used by the Luo tribe of Kenya to treat diarrhea while in Botswana, the black seeded landraces have been used to treat impotency (Udeh et al., 2020). In South Africa, nausea was controlled in pregnant women who chewed and swallowed the seed (Jideani and Jideani, 2021a; Khan et al., 2021). In Ghana, pounded Bambara seeds have been used to treat skin rashes and the powder mixed with the meat of guinea fowl have been used as a treatment against diarrhea in children (Akpalu et al., 2013).
Bambara Groundnut–Bacterial Interactions
The complexity of the rhizosphere is apparent in its role in plant health (Figure 2). Plants maintain this complex environment by secreting exudates into the rhizosphere (Olanrewaju et al., 2019). Plants secrete up to 40% of photosynthates that have access to roots in the rhizosphere (Berendsen et al., 2012). Because most of the soils are carbon deficient, these hot spots of carbon increase the microbial densities from 10 to 1,000 times, compared to bulk soil (Smalla et al., 2006). The increased concentration of microorganisms in the rhizosphere is due to the exchange of nutrients between the plant and the different taxa surrounding the root, which allows different types of associations. A number of factors influence the quantity and quality of root exudates, including plant species (Dennis et al., 2010), soil type (Berg and Smalla, 2009), plant developmental stage (Anzuay et al., 2021), pathogen attack (Olanrewaju et al., 2019), drought and heat stress (Ojuederie et al., 2019), and plant nutritional status (Carvalhais et al., 2011). If specific elements associated with the release of such exudates are better understood, novel approaches to enhancing beneficial microbial communities could be proposed.
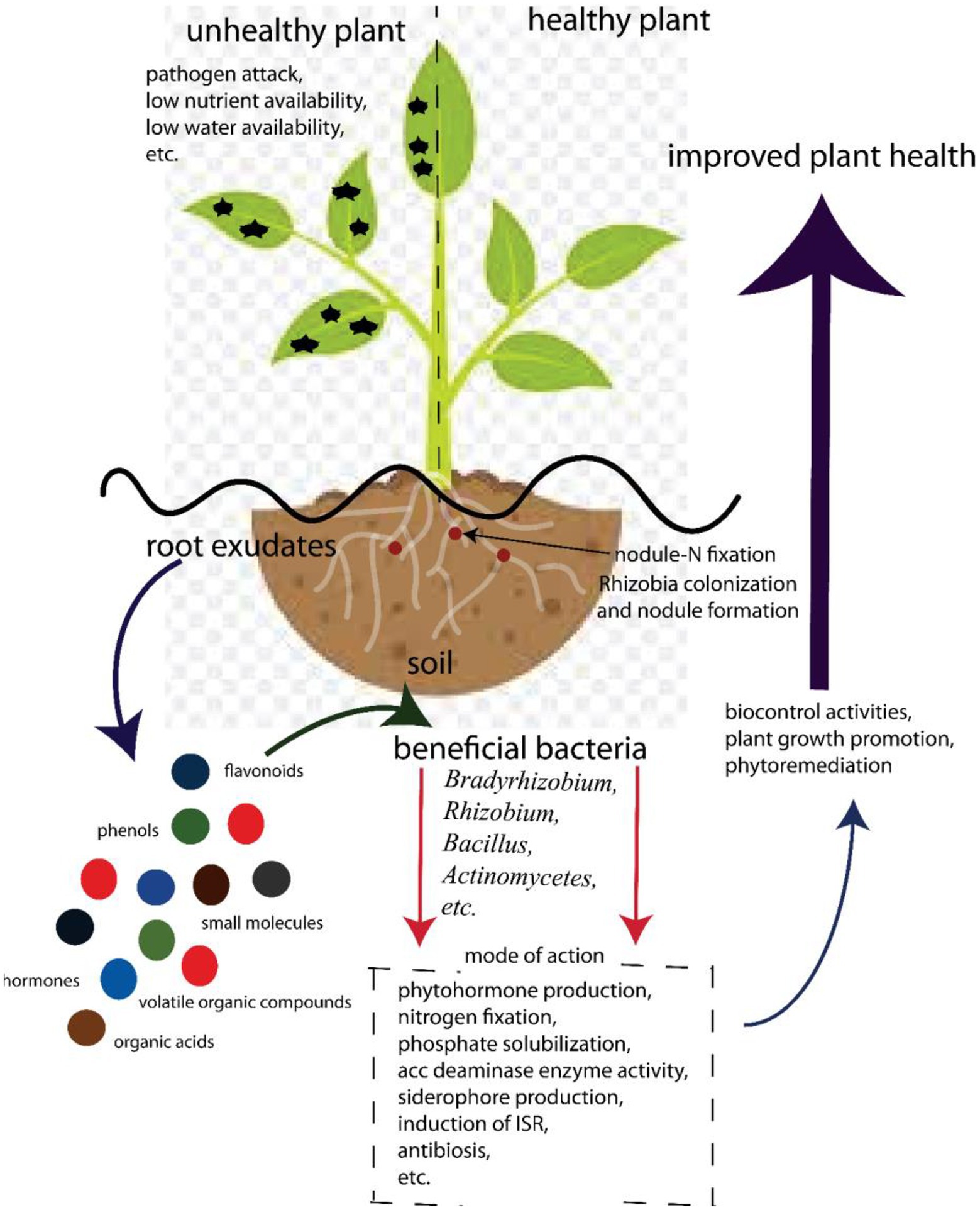
Figure 2. Beneficial associations involved in Bambara groundnut–bacterial interaction for food security.
Exudates released by plants contain phenolics, sugars, organic acids, and amino acids that attract both beneficial and non-beneficial microbes to the roots and serve as a source of carbon for these microbes (Figure 2). In exchange, beneficial microbes protect the plant against pathogens through well-documented mechanisms (Ahemad and Kibret, 2014; Olanrewaju et al., 2017; Backer et al., 2018). On the other hand, these carbon-containing compounds can also attract pathogens. Hence, they compete for nutrients, infect plants, and play a major role in structuring the rhizobiome.
Some plant species have been demonstrated to host-specific communities and attract protective microorganisms to suppress pathogens in the rhizosphere (Weinstein et al., 2021). Soil physical, chemical, and biological properties play an important role in the establishment of such plant–microbe interactions (Berendsen et al., 2012). Although pathogens can severely affect plant health, certain beneficial bacteria and fungi that also thrive in the rhizosphere, or inside plant tissues (endophytes) compete with these pathogens for space and nutrients; therefore exerting an antagonistic effect on them (Nihorimbere et al., 2011; Fadiji and Babalola, 2020; Verma et al., 2021).
PGPR grows in/on/or around root plant tissue and enhances plant growth, increases yield, protects plants against pathogens, and/or reduces biotic and abiotic stress (Trivedi et al., 2020; Ojuederie et al., 2021; Olanrewaju et al., 2021a). Growth promotion can be achieved directly by the interaction between the microbe and the host, as well as indirectly, due to antagonistic activities against plant pathogens. Direct interaction involves the production of phytohormones, which have been shown to inhibit or promote root growth, protect plants against biotic or abiotic stress, and improve nutrient acquisition by roots (Olanrewaju et al., 2017) and other direct-eliciting mechanisms. Because PGPR has the potential to replace chemical fertilizers and pesticides, it represents an environmentally sustainable alternative to increasing crop production and plant health. An interesting example of the role of microbial communities in plant nutrition and health is the interaction between rhizospheric fluorescent Pseudomonas and plants. Plants reduce soil iron (Fe) availability by acquiring iron and releasing exudates which attract the rhizospheric microbes that also utilize Fe. In Fe-stressed environments, siderophore-producing bacterial populations are enriched, which then suppress pathogens, such as fungi, for example, Oomycetes through competition for Fe. The plants, however, are able to utilize siderophores-bound iron, which enhances their growth (Lemanceau et al., 2013). This implies that the siderophore-producing microbes are able to produce the needed amount of Fe to the plants. Fe is an integral part of chlorophyll which is an important molecule in photosynthesis.
Another instance applied to plant disease suppression is the ability of resident microbiota in suppressive soils or compost to prevent pathogen infection (Hadar and Papadopoulou, 2012). In a soil suppressive to the fungal pathogen Rhizoctonia solani, Proteobacteria, Firmicutes, and Actinobacteria were prominent taxa found to be involved in disease suppression (Yin et al., 2013). There is also evidence to suggest that plants may use microbial communities to their own benefit to avoid infections (Mendes et al., 2011).
Symbiotic Interactions in Bambara Groundnut Rhizosphere
Rhizobia species (Rhizobium, Bradyrhizobium, Azorhizobium, Allorhizobium, Sinorhizobium, and Mesorhizobium) have been known to suppress the growth of plant pathogens and form nodules in symbiotic relationships with legumes (Babalola et al., 2017; Grönemeyer and Reinhold-Hurek, 2018; Ibny et al., 2019). The symbiotic relationship also results in the production of nitrogen-rich soil. BGN forms nodules and fixes nitrogen in partnership with Bradyrhizobium strains (Ibny et al., 2019). Nodule formation is important in Bambara–microbe interaction; this process starts with the production of compounds, such as betaines, flavonoids, and aldonic acid as root exudates from the plant (Figure 2). These compounds signal to the rhizobia in a compatible relationship with the compounds. This in turn enhances the production of the nod gene that induces nodulation by interacting with the nodD protein of the cell wall of the rhizobia (Wang et al., 2018; Walker et al., 2020). The rhizobia react to this inducement by producing and releasing the lipo-chito-oligosaccharide Nod factors, which bring about morphological changes in the root hair of the legume. This leads to the formation of an infection thread and the development of nodules that finally enhance fixation of nitrogen (Wang et al., 2018). Nod factors produced by rhizobia are important in plant growth as they promote germination of seeds and the development of seedlings (Kidaj et al., 2012).
Non-symbiotic Interactions in Bambara Groundnut Rhizosphere
Apart from the symbiotic relationship between plants and rhizobia, the production of phytohormones by these rhizobia, such as nod factors, riboflavin, and lipo-chito-oligosaccharide, can also stimulate plant growth and increase grain yield (Dakora, 2003). In addition, legumes generally produce phenolics (Tor-Roca et al., 2020) that help to suppress the activities of pathogens, make nutrients available to plants, and promote the growth of microorganisms with beneficial properties (Dakora, 2003).
Production of phenolic compounds mediates the production of nod gene inducers. The concentration of nod gene inducers regulate the production of nod factors around the root (Wang et al., 2018). When this is accumulated in the rhizosphere, it leads to biosynthesis of flavonoids, which also leads to an increased level of phytoalexin that is important for plant protection against pathogens (Dakora et al., 2015).
Rhizobia are also known to produce riboflavin (De Bruijn, 2015). It is a vitamin that is converted photochemically or by the actions of enzymes into lumichrome. This was evidenced in culture preparation from rhizobial cells. In its purified state, it was able to stimulate growth in maize, soybeans, and sorghum (Dakora et al., 2002). Rhizobia are important in suppressing the growth of pathogens of bacterial and fungal origins that infest sunflower, soybean, mungbean, and okra (Gopalakrishnan et al., 2015). Root exudates, such as phytosiderophores and organic acid anions, are important in making sure that minerals are available and circulate within the soil and agricultural systems (Dakora, 2003) that is the reason they are important in mixed cropping. Studies have shown that there was a continuous increase in yield and quality of seeds when BGN was inoculated with local strains of Bradyrhizobia. This is as a result of the increase in symbiotic nitrogen fixation activity of these microbes (Laurette et al., 2015).
Potential of Genomics and Beneficial Microbes in BGN Production
Plants attract specific microbiomes from the soil and air to their various compartments including rhizosphere, endosphere, and phyllosphere. Intra- and inter-specific variations among plant species inferred significant differences in the microbiome composition of the different species. This indicates that host genetics play an important role in microbiome assembly and function. By the late 21st century, temperature simulation reports have predicted an increase and this will lead to reduction in yield of all crops (Anderson et al., 2020) including BGN. Advanced breeding for BGN yield improvement and climate-resilient trait is affected by low level of genetic diversity available for breeding programs. Genetic diversity of BGN has not been fully exploited nor used in breeding programs, and recent genomics advances have increased the possibility of characterizing large germplasm collections through genome-wide association analysis for the identification of single nucleotide polymorphism (SNP) markers which can further be used in genomics assisted breeding for improved traits at any season or BGN growth stage.
Application of genomics and beneficial bacterial in BGN breeding is a way forward to improve yield and quality traits of the crop especially with the expected climate change impact on the environment which is expected to affect crop production.
Genomics for Improved BGN Production
Different markers are used for the genetic relationship and linkage mapping in BGN (Aliyu et al., 2016; Chai et al., 2017; Ho et al., 2017). The apparent use of the crop large germplasm for in improved breeding programs has caught the interest of breeders and geneticist. The genetic linkage map of BGN consisted of 11 linkage groups which originated from 223 markers (Ho et al., 2017). From the result obtained, it was observed that the linkage map showed synteny with their close relatives; mung bean, adzuki bean, and common bean genomes (Ho et al., 2017).
The Genetic Resource Centre (GRC) of the International Institute of Tropical Agriculture (IITA), Ibadan has the world’s largest collection of BGN accessions. Analysis of yield and yield stability, drought tolerance, nutritional and antinutritional contents have been carried out on the collection (Atoyebi et al., 2017; Adegboyega et al., 2021; Olanrewaju et al., 2021b,c). According to Paliwal et al. (2020), preliminary analysis showed significant variation in all accessions. This implies that there is a wide repertoire of genetic diversity and a great opportunity for genetic improvement in the crop. Furthermore, SNP genotyping has been applied paving the way for QTL analysis using GWAS. The aim is to develop core collections for future breeding research. So far, no GWAS study has been reported on BGN, hence, the importance of incorporating genomics into breeding programs cannot be over emphasized for this crop.
Furthermore, functional genomics increasingly allows the possibility of discovering new biocontrol, biostimulation, and biofertilization genes. Therefore, there is increased expectation in the exploitation of these genes for improved plant microbiome engineering. There is need for the implementation of global meta-omics approaches in elucidating the complex network of interactions in the plant microbiome. Integrative and comprehensive approach that combines multi-omics approaches, genetic engineering tools, synthetic biology, computational biology, bio-nanotechnology, and smart agricultural practices should be explored for the improvement of BGN production.
Beneficial Microbes for Improved BGN Production
Beneficial effects [such as biotic and abiotic stress tolerance (Etesami and Maheshwari, 2018; Fazeli-Nasab and Sayyed, 2019; Ojuederie et al., 2019; Goswami and Suresh, 2020), increasing nutrient uptake (Dhawi et al., 2015; Turan et al., 2016; Olanrewaju et al., 2017; Abdiev et al., 2019), pathogen control (Olanrewaju and Babalola, 2019a,b; Castaldi et al., 2021; Dave et al., 2021), and growth promotion (Ajilogba et al., 2017; Valetti et al., 2018; Hakim et al., 2021; Olanrewaju et al., 2021a)] of bacterial on various plant hosts are well researched and very much understood. Ability to control and select for beneficial bacteria would proffer an avenue to improve these traits. However, impact of plant genotype on the bacterial microbiome must be well understood for proper implementation of the bacterial-traits-plant improvement. As said earlier, various species of BGN vary in their trait’s responses due to different large genetic diversities available in the BGN germplasm. Hence, mechanistic insight into specie-specific bacterial microbiome influence is needed for proper implementation of the bacterial microbiome in improving BGN production. Application of mechanistic insight of bacterial microbiome regulation on underutilized legumes like BGN is limited by the inability of reconciling decades of works on molecular plant microbe interactions with the diverse and constantly changing microbiota (Bulgarelli et al., 2012).
Furthermore, impact of microbe–microbe interactions on BGN must be considered when addressing the use of beneficial microbes on the plant. For example, beneficial bacteria are said to be beneficial because of the benefits they proffer on plants which include promotion of root growth by phytohormones, acquisition of iron by siderophores, nitrogen fixation, and degradation of ethylene through the action of ACC deaminase enzyme, and nodule formation (which is majorly on legumes). Considering the role played by rhizobium in forming nodules and fixing nitrogen for BGN, how does this affect the bacterial dynamics in BGN microbiome? How does BGN differentiate beneficial microbes from pathogens? What are the specific exudates specific for the various microbial species attracted to the plant’s biome? For example, auxin can suppress plant defense thereby enhancing plant’s susceptibility to pathogen attacks (Su et al., 2021) which is partly due to many pathogens also producing their own auxin and in the process alter host auxin biology (Chen et al., 2015). The feedback effects of the host processes on the microbiome and vice versa must be well defined to fully exploit the full potentials of beneficial bacteria in the plant biome.
The complexity of bacterial interactions clearly makes targeted plant breeding for microbiome engineering challenging. However, application of metagenomics and the discovery of microbial hubs allows for discovery of important beneficial bacteria that can serve as biomarkers for key traits at any stage of BGN growth. Absence or presence of such bacterial biomarkers associated with BGN can be utilized to establish beneficial bacterial community structures for the plant.
Genomics-Enabled Approaches for Understanding BGN–Bacteria System Complexity
Engineering beneficial microbiota to improve BGN breeding requires improved linkage of the microbiota to the different BGN varieties. High-throughput amplicon sequencing has been productive in this aspect by linking microbial diversity to function in improving plant health (Brown et al., 2020). However, to understand how the environment, plant developmental stages, soil type, stress factors, and plant genotypes affect microbiome colonization, comprehensive characterization of BGN microbiome is needed. Through amplicon sequencing, especially 16S for bacteria microbiome, deep analysis of the BGN bacterial microbiota composition and functions can be predicted. Further application of long read metagenomics data with high-read sequencing will give a better depth and insight into the complexity and functions of BGN bacterial microbiota. Hence both approaches incorporated with BGN genome sequencing can be employed in targeted approaches to determine BGN genes that are involved in structuring the microbiome.
Furthermore, increasing availability of proteomics, transcriptomics, metagenomics, and metabolomics data will improve the identification of interaction and co-occurrence networks in BGN microbiome, thereby enabling the rapid discovery of microbiome interactions. This will result in improved breeding for important traits (Figure 3).
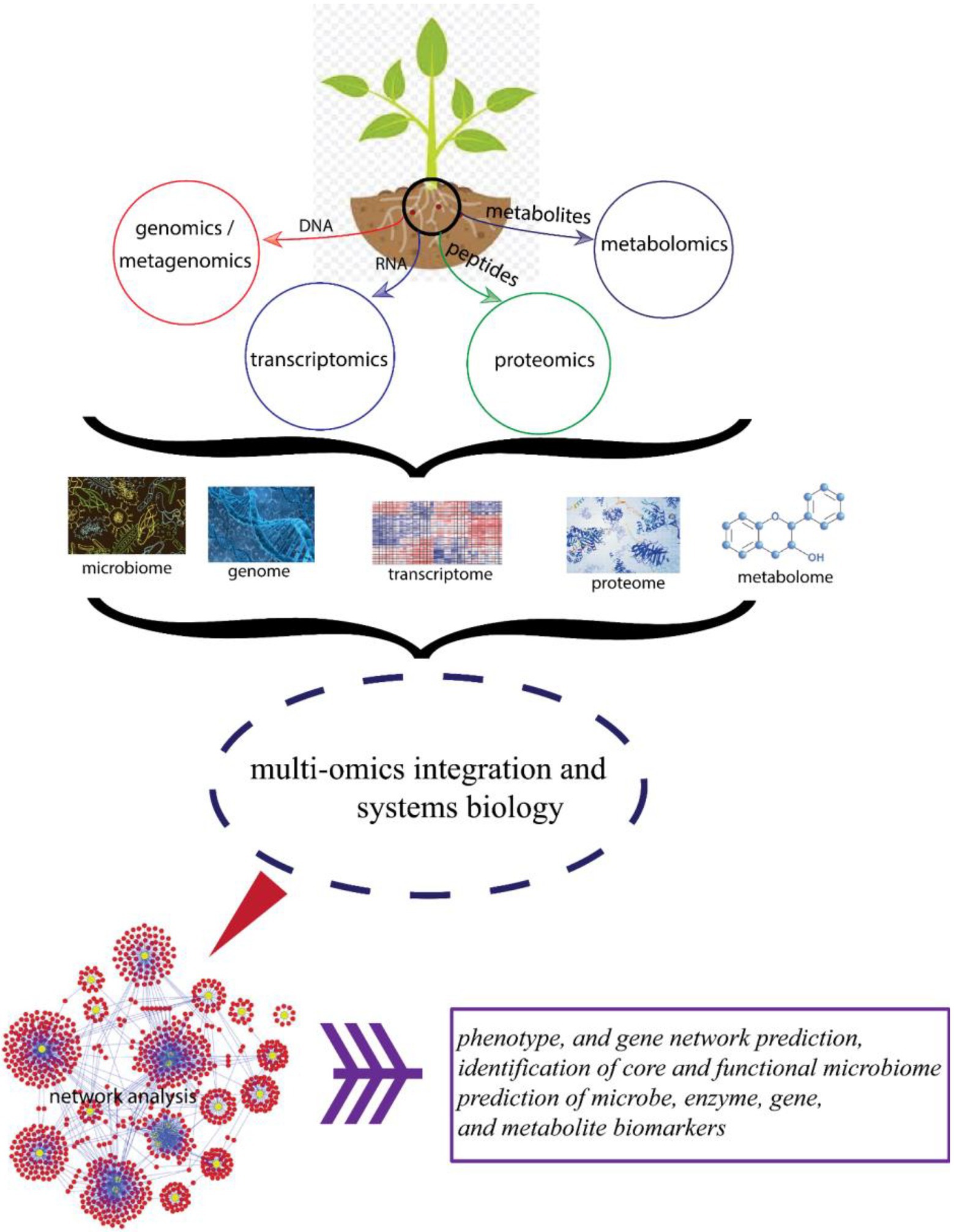
Figure 3. Schematic diagram showing different omics approaches and their beneficial effects toward improved plant health.
Transcriptomics for Studying BGN–Bacteria Associations
Transcriptomics investigates changes in expression of transcripts by using transcriptional regulators. Examples of such regulators include AhNFR5A, AhNFR5B, and NodD which regulate symbiotic processes and nodulation (Shu et al., 2020; Jiménez-Guerrero et al., 2021). In the expression of transcripts which occurs in response to microbe colonization, root exudates are key in these colonization activities. For example, flavonoids have been reported to be important in detecting protein-encoding genes in legumes during plant–microbe interactions in the process accounting for host specificity (Olanrewaju et al., 2019). Expression of protein-encoding genes in S. meliloti was regulated in M. sativa for the elucidation of gene interaction network (Chaudhary and Shukla, 2019).
The influence of plant growth-promoting microbes on the nodule and its bacteroid transcriptomes have been reported (Kelly et al., 2018; Lamouche et al., 2019). Researchers have employed laser-capture microdissection coupled with RNA sequencing for the total elucidation of expression patterns throughout the nodule compartment (Kusakin et al., 2021; Li et al., 2021). This technology is promising in fully understanding the gene expression patterns in nodules in response to microbe colonization.
Despite the availability of many studies addressing the transcriptional adaptations of microbes to their various hosts, many questions remain to be answered. For example, what is the molecular basis of host recognition? Specific role of signaling molecules, receptors, transcription factors, etc. Microbes might adapt their transcriptome in response to the host microbiome basically for survival in the biome. Therefore, does the transcriptomics modification of the microbiome by the expressing microbe have a causal effect on the genetic programming of the host? These and other yet unanswered questions may help in elucidating the mechanism of host and associated microbe transcriptomes for successful microbiome engineering application.
Comprehensive understanding of plant microbiome interactions will simplify the designing of microbial consortia capable of broad spectrum of plant growth-promoting abilities. Emerging technologies in microbiome functioning in addition to modern gene editing tools, such as CRISPR, RNAi, TALLEN, and so on, will make it possible to engineer microbial inoculants toward in situ microbiome engineering (Qiu et al., 2019; Trivedi et al., 2020).
Plants control diversity of their microbiome through a genetic network (Figure 3) in the process preventing dysbiosis. Hence, it is very possible with the availability of well-annotated genome, that BGN microbiome can be manipulated through genome editing of BGN plants with the recent advances in technologies, such as the CRISPR gene editing technology (Ramachandran and Bikard, 2019; Kumar and Dubey, 2020; Gao, 2021). With this, great opportunity lies ahead in comprehensive exploration of BGN–microbiome interactions to activate trait-specific members of its microbiota.
Proteomics for Studying BGN–Bacteria Association
Proteomics analysis technique improves our understanding of the regulation of protein expression. Studies at the proteome level can be used to identify cellular processes that occurs during interaction between plants and both beneficial bacteria as well as those induced by non-beneficial bacteria. In addition to identifying important proteins, proteomic studies can provide important information on genes that can be used in engineering plants toward biotic stress tolerance and improved plant growth. In simple organisms and complex organisms, proteomics has been a success in protein expression profiling. Proteomics was used to study plant–microbe interactions (Cooper et al., 2018), virulence factors in fungal pathogens (Rauwane et al., 2020), and plant’s responses to biotic and abiotic stress tolerance (Riviezzi et al., 2021). Proteomics has been applied in many studies involving legume–bacteria interactions (Table 1); however, no proteomic analysis has focused on BGN–bacteria association. This is partly due to the unavailability of a well-annotated reference genome.
The availability of a well-annotated BGN reference genome will facilitate better understanding of the molecular mechanisms of interactions between BGN and its microbiome. Further application of 15N/14N metabolic protein labeling, one-dimensional gel electrophoresis (1D), LC-electrospray ionization quantitative time-of-flight (ESI-Q-TOF), 2-D electrophoresis/MALDI-TOF, and shotgun proteomic approaches will aid identification of the high complexity of proteins from BGN and its associated bacteria which may yield significant advances in BGN microbiome studies. A comprehensive study on legume–rhizobium symbiosis has been reviewed by Khatabi et al. (2019).
Proteomics and Beneficial Bacteria in BGN–Microbe Interaction
The most abundant form of nitrogen; dinitrogen gas (N2) is inert and cannot be accessed by plants. However, it can be converted through nitrogen fixation process by nitrogen-fixing bacteria (Babalola et al., 2017). These nitrogen-fixing bacteria encode the nitrogenase enzyme which catalyzes the reduction of N2 to ammonia according to the following reaction:
Biological nitrogen fixation (BNF) is important in sustainable agriculture and mitigation of the environmental impacts posed by nitrogen fertilizers. Since the first report of BNF by plant-associated bacteria, there has been increased study of legume–bacteria symbiosis (Ferguson et al., 2019; Lindström and Mousavi, 2020; Schulte et al., 2021).
Glutamine synthetase, which uses ammonium as a substrate, catalyzes the conversion of glutamate to glutamine. This is followed by condensation of 2-oxoglutarate and glutamine to form glutamate. This process maintains glutamate homeostasis in nitrogen fixation. Hence, glutamine synthetase is believed to be of high importance in plant productivity with suggestions being made of it being a molecular marker for nitrogen uptake and utilization in plants (Swarbreck et al., 2011). Therefore, regulation of glutamine synthetase activities in BGN through proteomics could enhance knowledge for improved nitrogen efficiency and yield.
A symbiotic association for nodulation was found between the important model legume Medicago truncatula and Sinorhizobium meliloti (Larrainzar et al., 2007). In the study, proteomic approach was used to identify a total of 377 plant proteins in the nodules of M. truncatula using 2D-LC/MS/MS. Proteins reported are linked to nitrogen assimilation in nodules. Those involved in pathway of the synthesis of sulfur-containing amino acids were also reported. In another study by Nomura et al. (2010), the proteome of soybean-associating Bradyrhizobium japonicum was analyzed to evaluate time-dependent modulations in the proteome contents between 7 and 49 days of treatment.
Furthermore, recent advancement in visualization and modelling techniques allows identification of biomarker microbes in microbiome structure and function. Recent high-throughput culturing approaches can aid selection of significant microbes in microbial interactions which are related to better plant performance at various growth stages or in response to biotic and abiotic stresses. For example, Bez et al. (2022) identified several microbial groups showing negative correlation with Dickeya spp., a rice pathogen. These microbes can be targeted for improvements in controlling the pathogen. Applicable inoculants from these groups can be identified for specific and distinct abilities peculiar to each microbe in the group. Each microbe would exhibit distinct pathogen control mechanism and plant colonization; hence, large populations should be isolated and evaluated to obtain suitable microbes for further field trials.
In addition, gene expression analysis has aided determination of plant responses to bacteria colonization. However, the analyses of protein levels through proteomics technique are more essential for deciphering complex molecular mechanisms in plant–bacteria associations, especially as it applies to BGN–bacteria interactions.
Therefore, identification of other proteins involved in plant beneficial interactions through proteomics analysis of BGN–bacteria interaction is an avenue to harness the capacity of these beneficial bacteria on BGN yield. Furthermore, proteome studies not only provide insights into the activities of mRNA in the crosstalk between plants and beneficial bacteria, combined with metabolomics, it helps in elucidating the impact of the induced response on the plant phenotype.
Metabolomics for Studying BGN–Bacteria Associations
Metabolites are the end products of gene expressions. Metabolomics result directly correlates with the phenotype of the tissue or cell in study (Johnson et al., 2016; Figure 3). Hence, it is the link between genotypic effect on phenotypic responses. Root exudates in plants have been linked to the plant’s microbiome community. In Arabidopsis, for example, phenolic exudates positively correlates with the number of bacteria in the soil (Badri et al., 2013).
The mutualistic relationship between nitrogen-fixing rhizobia and soybean, an important legume, has been studied extensively. Soybeans’ dominant metabolites are of the classes isoflavones and saponins (Sugiyama, 2019), which act as signal molecules to attract nitrogen-fixing bacteria, such as Bradyrhizobium, Rhizobium, Mesorhizobium, and Azorhizobium. The importance of isoflavone in shaping the microbiome was revealed in the study by White et al. (2017) which showed the abundance of Xanthomonodaceae and Comamonodaceae in the rhizosphere of plants that can synthesize isoflavone over those that cannot.
Similarly, daidzein, an isoflavone, impacts the bacterial community in soybean root (Okutani et al., 2020). The findings in the study importantly report daidzein as specific in their recruitment of bacteria in the rhizosphere by attracting some and repelling others. Increased concentration of daidzein correlates with increased abundance of Comamonadaceae while rhizobia abundance decreases subsequently reducing the overall alpha diversity. This reduction in microbial diversity can be attributed to rhizobia not being able to efficiently use daidzein as a carbon source.
Pseudomonas sp. TLC6-6-5.4 and the endomycorrhizal mix of Glomus intraradices, Glomus mosseae, Glomus aggregatum, and Glomus etunicatum were inoculated in maize seedlings. After extraction and analysis of the metabolites by GC–MS (gas chromatography–mass spectrometry), upregulation of glyoxylate and dicarboxylate metabolism was observed (Dhawi et al., 2015). Single inoculation of PGPR alone or associated with endomycorrhizal mix increases the concentrations of mannitol, palmitic acid, lysine, stearic acid, and sucrose. Furthermore, metabolites produced showed correlation with nutrient uptake and biomass yield (Dhawi et al., 2015).
Metabolomics have been used to study mechanism of plant resistance to pathogens (Mhlongo et al., 2018; Zhu et al., 2018), drought stress (Mansour et al., 2021), production of antimicrobial compounds (Valette et al., 2020), and other focus which are presented in Table 1 as a result of inoculation with beneficial bacteria. These findings show that metabolomics is useful in screening BGN–bacteria associations for upregulation of important compounds that can be successfully regulated for BGN resistance to pathogens, drought tolerance, improved growth, yield, and nutrient components. Metabolomics, proteomics, and other omics applications will be useful in designing specific biofertilizer for BGN production.
Conclusion
Application of beneficial microbes holds great promise in sustainable agriculture and achieving food security; however, reported successes have varied largely because of inefficient/poor colonization, unstable environmental condition, limited persistence especially in the rhizosphere as result of high competition for nutrients. Genome engineering and root colonization with large populations of beneficial microbes may help provide a solution. Advances in synthetic biology enables engineering and improvement of non-model microbes toward specific trait improvement. Upon inoculation of these engineered microbes, follow up on the ability of these microbes for successful colonization of the plant microbiome and their efficacy for growth promotion, pathogen control, and improving crop yield in pilot plots, greenhouses, and field demonstrations should be explored before commercial adoption. Multi-environment trials over lengthy periods and seasons should be carried out to evaluate the environmental impacts on the field treatments.
Omics technologies have improved our understanding of mechanisms involved in major biological systems. The application of these techniques coupled with beneficial microbes will advance our understanding of specific microbial adaptions to BGN and improve our knowledge in engineering a beneficial microbiome to improve its production. Hence, employing multi-omics approach to study BGN–bacteria association will be fruitful in the coming years.
Author Contributions
All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
Funding
OB is deeply appreciative of the 7 years of National Research Foundation (NRF) incentive funding (UID81192).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We appreciate the National Research Foundation, South Africa, for grant that supports work in our laboratory.
References
Abdiev, A., Khaitov, B., Toderich, K., and Park, K. W. (2019). Growth, nutrient uptake and yield parameters of chickpea (Cicer arietinum L.) enhance by rhizobium and Azotobacter inoculations in saline soil. J. Plant Nutr. 42, 2703–2714. doi: 10.1080/01904167.2019.1655038
Adebola, M. I., Adelanwa, M. A., Mohammed, M. S., and Esson, A. E. (2017). Variation mineral compositions of some Bambara groundnut (Vigna subterranea (L.) Verdc.) accessions. Recent Res. Sci. Technol. 9, 06–09. doi: 10.25081/rrst.2017.9.3355
Adegboyega, T. T., Shoge, M. O., and Tamasi, A. A. (2021). Proximate and antinutrient composition of selected west African Bambara groundnut (Vigna subterranea (L.) Verdc.) accessions. J. Underutilized Leg. 3, 13–25.
Adeparusi, E., and Agbede, J. (2005). Evaluation of Leucaena and Gliricidia leaf protein concentrate as supplements to Bambara groundnut (Vigna subterranea (L.) Verdc.) in the diet of Oreochromis niloticus. Aquac. Nutr. 12, 335–342.
Agtuca, B. J., Stopka, S. A., Evans, S., Samarah, L., Liu, Y., Xu, D., et al. (2020). Metabolomic profiling of wild-type and mutant soybean root nodules using laser-ablation electrospray ionization mass spectrometry reveals altered metabolism. Plant J. 103, 1937–1958. doi: 10.1111/tpj.14815
Ahemad, M., and Kibret, M. (2014). Mechanisms and applications of plant growth promoting rhizobacteria: current perspective. J. King Saud Univ. Sci. 26, 1–20. doi: 10.1016/j.jksus.2013.05.001
Ajayi, F. A., and Lale, N. E. S. (2001). Susceptibility of unprotected seeds of local Bambara groundnut cultivars protected with insecticidal essential oils to infestation by Callosbrucuss maculates. J. Store. Product. 37, 47–62. doi: 10.1016/s0022-474x(00)00006-0
Ajilogba, C. F., Olanrewaju, O. S., and Babalola, O. O. (2017). “Application of bioinoculants for seed quality improvement,” in Microorganisms for Green Revolution. eds. Panpatte, D., Jhala, Y., Vyas, R., and Shelat, H. (Singapore: Springer), 265–280.
Akpalu, M., Atubilla, I., and Oppong-Sekyere, D. (2013). Assessing the level of cultivation and utilization of Bambara groundnut (Vigna subterranea (L.) Verdc.). In the Sumbrungu community of Bolgatanga, upper east region, Ghana. Int. J. Plant Ani. Environ. Sci. 3, 68–75.
Aliyu, S., Massawe, F., and Mayes, S. (2016). Genetic diversity and population structure of Bambara groundnut (Vigna subterranea (L.) Verdc.): synopsis of the past two decades of analysis and implications for crop improvement programmes. Genet. Resour. Crop. Evol. 63, 925–943. doi: 10.1007/s10722-016-0406-z
Anderson, R., Bayer, P. E., and Edwards, D. (2020). Climate change and the need for agricultural adaptation. Curr. Opin. Plant Biol. 56, 197–202. doi: 10.1016/j.pbi.2019.12.006
Anzuay, M. S., Viso, N. P., Ludueña, L. M., Morla, F. D., Angelini, J. G., and Taurian, T. (2021). Plant beneficial rhizobacteria community structure changes through developmental stages of peanut and maize. Rhizosphere 19:100407. doi: 10.1016/j.rhisph.2021.100407
Atoyebi, O. J., Osilesi, O., Adebawo, O., and Abberton, M. (2017). Evaluation of nutrient parameters of selected African accessions of Bambara groundnut (Vigna subterranea (L.) Verdc.). Ame. J. Food Nutr. 5, 83–89. doi: 10.12691/ajfn-5-3-1
Azam-Ali, S. N., Sesay, A., Karikari, S. K., Massawe, F. J., Aguilar-Manjarrez, J., Bannayan, M., et al. (2001). Assessing the potential of an underutilized crop–a case study using Bambara groundnut. Exp. Agric. 37, 433–472. doi: 10.1017/S0014479701000412
Babalola, O. O., Olanrewaju, O. S., Dias, T., Ajilogba, C. F., Kutu, F. R., and Cruz, C. (2017). “Biological nitrogen fixation: the role of underutilized leguminous plants,” in Microorganisms for Green Revolution. eds. Panpatte, D., Jhala, Y., Vyas, R., and Shelat, H. (Singapore: Springer), 431–443.
Backer, R., Rokem, J. S., Ilangumaran, G., Lamont, J., Praslickova, D., Ricci, E., et al. (2018). Plant growth-promoting rhizobacteria: context, mechanisms of action, and roadmap to commercialization of biostimulants for sustainable agriculture. Front. Plant Sci. 9:1473. doi: 10.3389/fpls.2018.01473
Badri, D. V., Chaparro, J. M., Zhang, R., Shen, Q., and Vivanco, J. M. (2013). Application of natural blends of phytochemicals derived from the root exudates of Arabidopsis to the soil reveal that phenolic-related compounds predominantly modulate the soil microbiome. J. Biol. Chem. 288, 4502–4512. doi: 10.1074/jbc.M112.433300
Bamishaiye, O., Adegbola, J., and Bamishaiye, E. (2011). Bambara groundnut: an under-utilized nut in Africa. Adv. Agric. Biotechnol. 1, 60–72.
Baudoin, J. P., and Mergeai, G. (eds.) (2001). Bambara groundnut. Brussels, Belgium: DGIC, Ministry of foreign affairs, External Trade and International development.
Berendsen, R. L., Pieterse, C. M., and Bakker, P. A. (2012). The rhizosphere microbiome and plant health. Trends Plant Sci. 17, 478–486. doi: 10.1016/j.tplants.2012.04.001
Berg, G., and Smalla, K. (2009). Plant species and soil type cooperatively shape the structure and function of microbial communities in the rhizosphere. FEMS Microbiol. Ecol. 68, 1–13. doi: 10.1111/j.1574-6941.2009.00654.x
Bez, C., Esposito, A., Thuy, H. D., Nguyen Hong, M., Valè, G., Licastro, D., et al. (2022). The rice foot rot pathogen Dickeya zeae alters the in-field plant microbiome. Environ. Microbiol. 23, 7671–7687. doi: 10.1111/1462-2920.15726
Brink, M., Ramolemana, G. M., and Sibuga, K. P. (2006). “Vigna subterranea (L.) Verdc. record from protabase,” in PROTA (Plant Resources of Tropical Africa/Ressources végétales de l’Afrique tropicale). eds. M. Brink and G. Belay (Wageningen, Netherlands: PROTA).
Brown, S. P., Grillo, M. A., Podowski, J. C., and Heath, K. D. (2020). Soil origin and plant genotype structure distinct microbiome compartments in the model legume Medicago truncatula. Microbiome 8:139. doi: 10.1186/s40168-020-00915-9
Bulgarelli, D., Rott, M., Schlaeppi, K., Van Themaat, E. V. L., Ahmadinejad, N., Assenza, F., et al. (2012). Revealing structure and assembly cues for Arabidopsis root-inhabiting bacterial microbiota. Nature 488, 91–95. doi: 10.1038/nature11336
Carvalhais, L. C., Dennis, P. G., Fedoseyenko, D., Hajirezaei, M. R., Borriss, R., and Von Wirén, N. (2011). Root exudation of sugars, amino acids, and organic acids by maize as affected by nitrogen, phosphorus, potassium, and iron deficiency. J. Plant Nutr. Soil Sci. 174, 3–11. doi: 10.1002/jpln.201000085
Castaldi, S., Petrillo, C., Donadio, G., Piaz, F. D., Cimmino, A., Masi, M., et al. (2021). Plant growth promotion function of Bacillus sp. strains isolated from salt-pan rhizosphere and their biocontrol potential against Macrophomina phaseolina. Int. J. Mol. Sci. 22:3324. doi: 10.3390/ijms22073324
Chai, H. H., Ho, W. K., Graham, N., May, S., Massawe, F., and Mayes, S. (2017). A cross-species gene expression marker-based genetic map and QTL analysis in Bambara groundnut. Genes 8:84. doi: 10.3390/genes8020084
Chaparro, J. M., Badri, D. V., and Vivanco, J. M. (2014). Rhizosphere microbiome assemblage is affected by plant development. ISME J. 8, 790–803. doi: 10.1038/ismej.2013.196
Chaudhary, T., and Shukla, P. (2019). Bioinoculant capability enhancement through metabolomics and systems biology approaches. Brief. Funct. Genomics 18, 159–168. doi: 10.1093/bfgp/elz011
Chen, Z., Cui, Q., Liang, C., Sun, L., Tian, J., and Liao, H. (2011). Identification of differentially expressed proteins in soybean nodules under phosphorus deficiency through proteomic analysis. Proteomics 11, 4648–4659. doi: 10.1002/pmic.201100231
Chen, Z., Hu, L., Han, N., Hu, J., Yang, Y., Xiang, T., et al. (2015). Overexpression of a miR393-resistant form of transport inhibitor response protein 1 (mTIR1) enhances salt tolerance by increased osmoregulation and Na+ exclusion in Arabidopsis thaliana. Plant Cell Physiol. 56, 73–83. doi: 10.1093/pcp/pcu149
Chen, L., Wu, Q., He, W., He, T., Wu, Q., and Miao, Y. (2019). Combined de novo transcriptome and metabolome analysis of common bean response to Fusarium oxysporum f. sp. phaseoli infection. Int. J. Mol. Sci. 20:6278. doi: 10.3390/ijms20246278
Cook, D. (2017). Small scale farmers utilization and perceptions of Bambara groundnut production in South Africa: a case study in a semi-arid region of Limpopo. Master of Philosophy Dissertation, University of Cape Town.
Cooper, B., Campbell, K. B., Beard, H. S., Garrett, W. M., Mowery, J., Bauchan, G. R., et al. (2018). A proteomic network for symbiotic nitrogen fixation efficiency in Bradyrhizobium elkanii. Mol. Plant-Microbe Interact. 31, 334–343. doi: 10.1094/MPMI-10-17-0243-R
Dakora, F. D. (2003). Defining new roles for plant and rhizobial molecules in sole and mixed plant cultures involving symbiotic legumes. New Phytol. 158, 39–49. doi: 10.1046/j.1469-8137.2003.00725.x
Dakora, F. D., Matiru, V. N., and Kanu, A. S. (2015). Rhizosphere ecology of lumichrome and riboflavin, two bacterial signal molecules eliciting developmental changes in plants. Front. Plant Sci. 6:700. doi: 10.3389/fpls.2015.00700
Dakora, F. D., Matiru, V., King, M., and Phillips, D. A. (2002). “Plant growth promotion in legumes and cereals by lumichrome, a rhizobial signal metabolite,” in Nitrogen Fixation: Global Perspectives. eds. T. M. Finan, M. R. O'brian, D. B. Layzell, K. Vessey, and W. E. Newton (Wallingford, UK: CABI Publishing).
Dave, K., Gothalwal, R., Singh, M., and Joshi, N. (2021). Facets of rhizospheric microflora in biocontrol of phytopathogen Macrophomina phaseolina in oil crop soybean. Arch. Microbiol. 203, 405–412. doi: 10.1007/s00203-020-02046-z
De Bruijn, F. J. (2015). “Biological nitrogen fixation,” in Principles of Plant-Microbe Interactions. ed. B. Lugtenberg (Switzerland: Springer), 215–224.
Dennis, P. G., Miller, A. J., and Hirsch, P. R. (2010). Are root exudates more important than other sources of rhizodeposits in structuring rhizosphere bacterial communities? FEMS Microbiol. Ecol. 72, 313–327. doi: 10.1111/j.1574-6941.2010.00860.x
Dhawi, F., Datta, R., and Ramakrishna, W. (2015). Mycorrhiza and PGPB modulate maize biomass, nutrient uptake and metabolic pathways in maize grown in mining-impacted soil. Plant Physiol. Biochem. 97, 390–399. doi: 10.1016/j.plaphy.2015.10.028
Durán, D., Albareda, M., García, C., Marina, A.-I., Ruiz-Argüeso, T., and Palacios, J.-M. (2021). Proteome analysis reveals a significant host-specific response in Rhizobium leguminosarum bv. Viciae endosymbiotic cells. Mol. Cell. Proteomics 20:100009. doi: 10.1074/mcp.RA120.002276
Emelike, N., and Barber, L. (2018). Assessment of food quality and the associated mycoflora of okpa, a local recipe from Bambara groundnut. Res. J. Food Sci. Quality Control 4, 1–5.
Etesami, H., and Maheshwari, D. K. (2018). Use of plant growth promoting rhizobacteria (PGPRs) with multiple plant growth promoting traits in stress agriculture: action mechanisms and future prospects. Ecotoxicol. Environ. Saf. 156, 225–246. doi: 10.1016/j.ecoenv.2018.03.013
Fadiji, A. E., and Babalola, O. O. (2020). Elucidating mechanisms of endophytes used in plant protection and other bioactivities with multifunctional prospects. Front. Bioeng. Biotechnol. 8:467. doi: 10.3389/fbioe.2020.00467
Fazeli-Nasab, B., and Sayyed, R. (2019). “Plant growth-promoting rhizobacteria and salinity stress: a journey into the soil,” in Plant Growth Promoting Rhizobacteria for Sustainable Stress Management. eds. Sayyed, R., Arora, N., and Reddy, M. (Singapore: Springer), 21–34.
Ferguson, B. J., Minamisawa, K., Muñoz, N. B., and Lam, H.-M. (2019). Metabolic adjustments and gene expression reprogramming for symbiotic nitrogen fixation in legume nodules. Front. Plant Sci. 10:898. doi: 10.3389/fpls.2019.00898
Foyer, C. H., Lam, H.-M., Nguyen, H. T., Siddique, K. H. M., Varshney, R. K., Colmer, T. D., et al. (2016). Neglecting legumes has compromised human health and sustainable food production. Nat. Plants 2:16112. doi: 10.1038/nplants.2016.112
Gao, C. (2021). Genome engineering for crop improvement and future agriculture. Cell 184, 1621–1635. doi: 10.1016/j.cell.2021.01.005
Gopalakrishnan, S., Sathya, A., Vijayabharathi, R., Varshney, R. K., Gowda, C. L., and Krishnamurthy, L. (2015). Plant growth promoting rhizobia: challenges and opportunities. 3 Biotech 5, 355–377. doi: 10.1007/s13205-014-0241-x
Goswami, M., and Suresh, D. (2020). Plant growth-promoting rhizobacteria—alleviators of abiotic stresses in soil: a review. Pedosphere 30, 40–61. doi: 10.1016/S1002-0160(19)60839-8
Grönemeyer, J. L., and Reinhold-Hurek, B. (2018). Diversity of Bradyrhizobia in sub-Sahara Africa: a rich resource. Front. Microbiol. 9:2194. doi: 10.3389/fmicb.2018.02194
Hadar, Y., and Papadopoulou, K. K. (2012). Suppressive composts: microbial ecology links between abiotic environments and healthy plants. Annu. Rev. Phytopathol. 50, 133–153. doi: 10.1146/annurev-phyto-081211-172914
Haichar, F.-E. Z., Santaella, C., Heulin, T., and Achouak, W. (2014). Root exudates mediated interactions belowground. Soil Biol. Biochem. 77, 69–80. doi: 10.1016/j.soilbio.2014.06.017
Hakim, S., Naqqash, T., Nawaz, M. S., Laraib, I., Siddique, M. J., Zia, R., et al. (2021). Rhizosphere engineering with plant growth-promoting microorganisms for agriculture and ecological sustainability. Front. Sust. Food Sys. 5:617157. doi: 10.3389/fsufs.2021.617157
Halimi, R. A., Barkla, B. J., Mayes, S., and King, G. J. (2019). The potential of the underutilized pulse Bambara groundnut (Vigna subterranea (L.) Verdc.) for nutritional food security. J. Food Compos. Anal. 77, 47–59. doi: 10.1016/j.jfca.2018.12.008
Hassen, A. I. I., Van Vuuren, A., Bopape, F. L., and Gerrano, A. S. (2022). Nodulation compatibility and symbiotic performance of rhizobia spp. with different landraces of Bambara groundnut (Vigna Subterranea (L.) Verdc.) collections. Research Square 1–23 [Preprint]. doi: 10.21203/rs.3.rs-1233082/v1
Hillocks, R., Bennett, C., and Mponda, O. (2012). Bambara nut: a review of utilisation, market potential and crop improvement. Afr. Crop. Sci. J. 20, 1–16.
Hlanga, N. C., Modi, A. T., and Mathew, I. (2021). Evaluating nutritional content among Bambara groundnut lines. J. Food Compos. Anal. 102:104053. doi: 10.1016/j.jfca.2021.104053
Ho, W. K., Chai, H. H., Kendabie, P., Ahmad, N. S., Jani, J., Massawe, F., et al. (2017). Integrating genetic maps in Bambara groundnut [Vigna subterranea (L) Verdc.] and their syntenic relationships among closely related legumes. BMC Genomics 18:91. doi: 10.1186/s12864-016-3393-8
Ibny, F. Y., Jaiswal, S. K., Mohammed, M., and Dakora, F. D. (2019). Symbiotic effectiveness and ecologically adaptive traits of native rhizobial symbionts of Bambara groundnut (Vigna subterranea L. Verdc.) in Africa and their relationship with phylogeny. Sci. Rep. 9:12666. doi: 10.1038/s41598-019-48944-1
Ijarotimi, S. O., and Esho, R. T. (2009). Comparison of nutritional composition and anti-nutrient status of fermented, germinated and roasted Bambara groundnut seeds (Vigna subterranea). British Food J. 111, 376–386. doi: 10.1108/00070700910951515
Jideani, V. A., and Diedericks, C. F. (2014). “Nutritional, therapeutic, and prophylactic properties of Vigna subterranea and Moringa oleifera,” in Antioxidant-Antidiabetic Agents and Human Health. ed. Oguntibeju, O. (Croatia: InTech), 187–207.
Jideani, V. A., and Jideani, A. I. (2021a). “Ethnonutritional and ethnomedicinal uses of Bambara groundnut,” in Bambara Groundnut: Utilization and Future Prospects (Switzerland: Springer), 49–60.
Jideani, V. A., and Jideani, A. I. (2021b). “World production, ecology and postharvest handling of Bambara groundnut,” in Bambara Groundnut: Utilization and Future Prospects (Switzerland: Springer), 11–29.
Jiménez-Guerrero, I., Moreno-De Castro, N., and Pérez-Montaño, F. (2021). One door closes, another opens: when nodulation impairment with natural hosts extends rhizobial host-range. Environ. Microbiol. 23, 1837–1841. doi: 10.1111/1462-2920.15353
Johnson, C. H., Ivanisevic, J., and Siuzdak, G. (2016). Metabolomics: beyond biomarkers and towards mechanisms. Nat. Rev. Mol. Cell Biol. 17:451. doi: 10.1038/nrm.2016.25
Kang, W., Jiang, Z., Chen, Y., Wu, F., Liu, C., Wang, H., et al. (2020). Plant transcriptome analysis reveals specific molecular interactions between alfalfa and its rhizobial symbionts below the species level. BMC Plant Biol. 20:293. doi: 10.1186/s12870-020-02503-3
Karmakar, K., Kundu, A., Rizvi, A. Z., Dubois, E., Severac, D., Czernic, P., et al. (2019). Transcriptomic analysis with the progress of symbiosis in ‘crack-entry’ legume Arachis hypogaea highlights its contrast with ‘infection thread’ adapted legumes. Mol. Plant-Microbe Interact. 32, 271–285. doi: 10.1094/MPMI-06-18-0174-R
Kelly, S., Mun, T., Stougaard, J., Ben, C., and Andersen, S. U. (2018). Distinct Lotus japonicus transcriptomic responses to a spectrum of bacteria ranging from symbiotic to pathogenic. Front. Plant Sci. 9:1218. doi: 10.3389/fpls.2018.01218
Khan, M. M. H., Rafii, M. Y., Ramlee, S. I., Jusoh, M., and Al-Mamun, M. (2021). Bambara groundnut (Vigna subterranea L. Verdc): A crop for the new millennium, its genetic diversity, and improvements to mitigate future food and nutritional challenges. Sustainability 13:5530. doi: 10.3390/su13105530
Khan, M. M. H., Rafii, M. Y., Ramlee, S. I., Jusoh, M., and Mamun, A. (2020). Genetic variability, heritability, and clustering pattern exploration of Bambara groundnut (Vigna subterranea L. Verdc) accessions for the perfection of yield and yield-related traits. Biomed Res. Int. 2020:2195797. doi: 10.1155/2020/2195797
Khatabi, B., Gharechahi, J., Ghaffari, M. R., Liu, D., Haynes, P. A., Mckay, M. J., et al. (2019). Plant–microbe symbiosis: what has proteomics taught us? Proteomics 19:1800105. doi: 10.1002/pmic.201800105
Kidaj, D., Wielbo, J., and Skorupska, A. (2012). Nod factors stimulate seed germination and promote growth and nodulation of pea and vetch under competitive conditions. Microbiol. Res. 167, 144–150. doi: 10.1016/j.micres.2011.06.001
Kumar, A., and Dubey, A. (2020). Rhizosphere microbiome: engineering bacterial competitiveness for enhancing crop production. J. Adv. Res. 24, 337–352. doi: 10.1016/j.jare.2020.04.014
Kumar, S., Paritosh, K., Pareek, N., Chawade, A., and Vivekanand, V. (2018). De-construction of major Indian cereal crop residues through chemical pretreatment for improved biogas production: an overview. Renew. Sust. Energ. Rev. 90, 160–170. doi: 10.1016/j.rser.2018.03.049
Kusakin, P. G., Serova, T. A., Gogoleva, N. E., Gogolev, Y. V., and Tsyganov, V. E. (2021). Laser microdissection of Pisum sativum L. nodules followed by RNA-Seq analysis revealed crucial transcriptomic changes during infected cell differentiation. Agronomy 11:2504. doi: 10.3390/agronomy11122504
Lamouche, F., Chaumeret, A., Guefrachi, I., Barrière, Q., Pierre, O., Guérard, F., et al. (2019). From intracellular bacteria to differentiated bacteroids: transcriptome and metabolome analysis in Aeschynomene nodules using the Bradyrhizobium sp. strain ORS285 bclA mutant. J. Bacteriol. 201:e00191–19. doi: 10.1128/JB.00191-19
Laplaze, L., Sparvoli, F., Masmoudi, K., and Hash, C. T. (2018). Harvesting plant and microbial biodiversity for sustainably enhanced food security. Front. Plant Sci. 9:42. doi: 10.3389/fpls.2018.00042
Larrainzar, E. B., Wienkoop, S., Weckwerth, W., Ladrera, R. N., Arrese-Igor, C., and GonzáLez, E. M. (2007). Medicago truncatula root nodule proteome analysis reveals differential plant and bacteroid responses to drought stress. Plant Physiol. 144, 1495–1507. doi: 10.1104/pp.107.101618
Laurette, N. N., Maxémilienne, N. B., Henri, F., Souleymanou, A., Kamdem, K., Albert, N., et al. (2015). Isolation and screening of indigenous Bambara groundnut (Vigna Subterranea) nodulating bacteria for their tolerance to some environmental stresses. Ame. J. Microbiol. Res. 3, 65–75. doi: 10.12691/ajmr-3-2-5
Lemanceau, P., Mazurier, S., Avoscan, L., Robin, A., and Briat, J. F. (2013). “Reciprocal interactions between plants and fluorescent pseudomonas in relation to iron in the rhizosphere,” in Reciprocal Interactions between Plants and Fluorescent Pseudomonas in Relation to Iron in the Rhizosphere. ed. F. J. De Bruijn (Hoboken, NJ, USA: Wiley-Blackwell), 1181–1189.
Li, H., Schilderink, S., Cao, Q., Kulikova, O., and Bisseling, T. (2021). Plant-specific histone deacetylases are essential for early and late stages of Medicago nodule development. Plant Physiol. 186, 1591–1605. doi: 10.1093/plphys/kiab140
Lindström, K., and Mousavi, S. A. (2020). Effectiveness of nitrogen fixation in rhizobia. Microb. Biotechnol. 13, 1314–1335. doi: 10.1111/1751-7915.13517
Majola, N. G., Gerrano, A. S., and Shimelis, H. (2021). Bambara groundnut (Vigna subterranea [L.] Verdc.) production, utilisation and genetic improvement in sub-Saharan Africa. Agronomy 11:1345. doi: 10.3390/agronomy11071345
Mamontova, T., Afonin, A. M., Ihling, C., Soboleva, A., Lukasheva, E., Sulima, A. S., et al. (2019). Profiling of seed proteome in Pea (Pisum sativum L.) lines characterized with high and low responsivity to combined inoculation with nodule bacteria and Arbuscular Mycorrhizal fungi. Molecules 24:1603. doi: 10.3390/molecules24081603
Mansour, E., Mahgoub, H. A., Mahgoub, S. A., El-Sobky, E.-S. E., Abdul-Hamid, M. I., Kamara, M. M., et al. (2021). Enhancement of drought tolerance in diverse Vicia faba cultivars by inoculation with plant growth-promoting rhizobacteria under newly reclaimed soil conditions. Sci. Rep. 11:24142. doi: 10.1038/s41598-021-02847-2
Masindeni, D. R. (2006). Evaluation of Bambara groundnut (Vigna subterranea) for yield stability and yield related characteristics. MSc dissertation M.Sc. Thesis, University of the Free State.
Massawe, F., Mayes, S., and Cheng, A. (2016). Crop diversity: An unexploited treasure trove for food security. Trends Plant Sci. 21, 365–368. doi: 10.1016/j.tplants.2016.02.006
Mayes, S., Ho, W. K., Chai, H. H., Gao, X., Kundy, A. C., Mateva, K. I., et al. (2019). Bambara groundnut: an exemplar underutilised legume for resilience under climate change. Planta 250, 803–820. doi: 10.1007/s00425-019-03191-6
Mazahib, A., Nuha, M., Salawa, I., and Babiker, E. (2013). Some nutritional attributes of Bambara groundnut as influenced by domestic processing. Int. Food Res. J. 20, 1165–1171.
Mendes, R., Kruijt, M., De Bruijn, I., Dekkers, E., Van Der Voort, M., Schneider, J. H., et al. (2011). Deciphering the rhizosphere microbiome for disease-suppressive bacteria. Science 332, 1097–1100. doi: 10.1126/science.1203980
Mhlongo, M. I., Piater, L. A., Madala, N. E., Labuschagne, N., and Dubery, I. A. (2018). The chemistry of plant–microbe interactions in the rhizosphere and the potential for metabolomics to reveal signaling related to defense priming and induced systemic resistance. Front. Plant Sci. 9:112. doi: 10.3389/fpls.2018.00112
Mkandawire, C. H. (2007). Review of Bambara groundnut (Vigna subterranea (L.) Verdc.) production in sub-Sahara Africa. Agric. J. 2, 464–470.
Mubaiwa, J., Fogliano, V., Chidewe, C., Bakker, E. J., and Linnemann, A. R. (2018). Utilization of Bambara groundnut (Vigna subterranea (L.) Verdc.) for sustainable food and nutrition security in semi-arid regions of Zimbabwe. PLoS One 13:e0204817. doi: 10.1371/journal.pone.0204817
Murevanhema, Y. Y., and Jideani, V. A. (2013). Potential of Bambara groundnut (Vigna subterranea (L.) Verdc) milk as a probiotic beverage—A review. Crit. Rev. Food Sci. Nutr. 53, 954–967. doi: 10.1080/10408398.2011.574803
Nihorimbere, V., Ongena, M., Smargiassi, M., and Thonart, P. (2011). Beneficial effect of the rhizosphere microbial community for plant growth and health. Biotechnol. Agron. Soc. Environ. 15, 327–337.
Nomura, M., Arunothayanan, H., Van Dao, T., Le, H. T. P., Kaneko, T., Sato, S., et al. (2010). Differential protein profiles of Bradyrhizobium japonicum USDA110 bacteroid during soybean nodule development. Soil Sci. Plant Nutr. 56, 579–590. doi: 10.1111/j.1747-0765.2010.00500.x
Nyau, V., Mwelwa-Zgambo, L., Chirwa-Moonga, T., Nthani, D., Prakash, S., Rodrigues, J., et al. (2020). Some nutritional and physical properties of different Zambian market classes of Bambara groundnut (Vigna subterranea). J. Food Res. 9, 34–40. doi: 10.5539/jfr.v9n1p34
Ojiewo, C., Keatinge, D. J. D. H., Hughes, J., Tenkouano, A., Nair, R., Varshney, R., et al. (2015). The role of vegetables and legumes in assuring food, nutrition, and income security for vulnerable groups in sub-Saharan Africa. World Med. Health Policy 7, 187–210. doi: 10.1002/wmh3.148
Ojuederie, O. B., Chukwuneme, C. F., Samuel, O., Olanrewaju, M. A., Adegboyega, T. T., and Babalola, O. O. (2021). “Contribution of microbial inoculants in sustainable maintenance of human health, including test methods and evaluation of safety of microbial pesticide microorganisms,” in Biopesticides: Botanicals and Microorganisms for Improving Agriculture and Human Health. eds. T. O. Adejumo and R. T. Vogele (Berlin, Germany: Logos Verlag Berlin GmbH), 215.
Ojuederie, O. B., Olanrewaju, O. S., and Babalola, O. O. (2019). Plant growth promoting rhizobacterial mitigation of drought stress in crop plants: implications for sustainable agriculture. Agronomy 9:712. doi: 10.3390/agronomy9110712
Okutani, F., Hamamoto, S., Aoki, Y., Nakayasu, M., Nihei, N., Nishimura, T., et al. (2020). Rhizosphere modelling reveals spatiotemporal distribution of daidzein shaping soybean rhizosphere bacterial community. Plant Cell Environ. 43, 1036–1046. doi: 10.1111/pce.13708
Olanrewaju, O. S., Ayangbenro, A. S., Glick, B. R., and Babalola, O. O. (2019). Plant health: feedback effect of root exudates-rhizobiome interactions. Appl. Microbiol. Biotechnol. 103, 1155–1166. doi: 10.1007/s00253-018-9556-6
Olanrewaju, O. S., Ayilara, M. S., Ayangbenro, A. S., and Babalola, O. O. (2021a). Genome mining of three plant growth-promoting bacillus species from maize rhizosphere. Appl. Biochem. Biotechnol. 193, 3949–3969. doi: 10.1007/s12010-021-03660-3
Olanrewaju, O. S., and Babalola, O. O. (2019a). Bacterial consortium for improved Maize (Zea mays L.) production. Microorganisms 7:519. doi: 10.3390/microorganisms7110519
Olanrewaju, O. S., and Babalola, O. O. (2019b). Streptomyces: implications and interactions in plant growth promotion. Appl. Microbiol. Biotechnol. 103, 1179–1188. doi: 10.1007/s00253-018-09577-y
Olanrewaju, O. S., Glick, B. R., and Babalola, O. O. (2017). Mechanisms of action of plant growth promoting bacteria. World J. Microbiol. Biotechnol. 33:197. doi: 10.1007/s11274-017-2364-9
Olanrewaju, O. S., Oyatomi, O., Babalola, O. O., and Abberton, M. (2021b). Genetic diversity and environmental influence on growth and yield parameters of Bambara groundnut. Front. Plant Sci. 12:796352. doi: 10.3389/fpls.2021.796352
Olanrewaju, O. S., Oyatomi, O., Babalola, O. O., and Abberton, M. (2021c). GGE biplot analysis of genotype X environment interaction and yield stability in Bambara groundnut. Agronomy 11:1839. doi: 10.3390/agronomy11091839
Olanrewaju, O. S., Oyatomi, O., Babalola, O. O., and Abberton, M. (2022). Breeding potentials of Bambara groundnut for food and nutrition security in the face of climate change. Front. Plant Sci. 12:798993. doi: 10.3389/fpls.2021.798993
Olukolu, B. A., Mayes, S., Stadler, F., Quat Ng, N., Fawole, I., Dominique, D., et al. (2012). Genetic diversity in Bambara groundnut (Vigna subterranea (L.) Verdc.) as revealed by phenotypic descriptors and DArT marker analysis. Genet. Resour. Crop. Evol. 59, 347–358. doi: 10.1007/s10722-011-9686-5
Oluwole, O. B., Nicholas-Okpara, V. A. N., Elemo, G., Adeyoju, O., Ibekwe, D., and Adegboyega, M. O. (2021). Medicinal uses, nutraceutical potentials and traditional farm production of Bambara beans and Pigeon pea. Global J. Epidemiol. Public Health 6, 41–50. doi: 10.12974/2313-0946.2021.06.01.3
Omoikhoje, S. O. (2008). Assessment of the nutritive value of Bambara groundnut as influenced by cooking time. Livest. Res. Rural. Dev. 20:55.
Onwubiko, N., Uguru, M., Ngwuta, A., Inyang, E., and Nnajiemere, O. (2011). Floral biology of Bambara groundnut [Vigna subterranea (L.) Verdc.]. J. Plant Breed. Crop Sci. 3, 293–295.
Opoku, M. K. (2010). Response of Bambara groundnut landraces to heat and drought stress in the Guinea Savanna and transition zones of Ghana. Master Of Science (Agronomy), Kwame Nkrumah University Of Science and Technology.
Paliwal, R., Abberton, M., Faloye, B., and Olaniyi, O. (2020). Developing the role of legumes in West Africa under climate change. Curr. Opin. Plant Biol. 56, 242–258. doi: 10.1016/j.pbi.2020.05.002
Pui, L.-P., Tan, W.-C., Kong, I., and Tan, C.-H. (2021). Drought-tolerant Bambara groundnuts as future food: a comprehensive review of its properties and applications in food. British Food J. 1–15. doi: 10.1108/BFJ-04-2021-0436, [Epub ahead of print]
Qiu, Z., Egidi, E., Liu, H., Kaur, S., and Singh, B. K. (2019). New frontiers in agriculture productivity: optimised microbial inoculants and in situ microbiome engineering. Biotechnol. Adv. 37:107371. doi: 10.1016/j.biotechadv.2019.03.010
Ramachandran, G., and Bikard, D. (2019). Editing the microbiome the CRISPR way. Philos. Trans. R. Soc. B 374:20180103. doi: 10.1098/rstb.2018.0103
Ramírez-Ordorica, A., Valencia-Cantero, E., Flores-Cortez, I., Carrillo-Rayas, M. T., Elizarraraz-Anaya, M. I. C., Montero-Vargas, J., et al. (2020). Metabolomic effects of the colonization of Medicago truncatula by the facultative endophyte Arthrobacter agilis UMCV2 in a foliar inoculation system. Sci. Rep. 10:8426. doi: 10.1038/s41598-020-65314-4
Rauwane, M. E., Ogugua, U. V., Kalu, C. M., Ledwaba, L. K., Woldesemayat, A. A., and Ntushelo, K. (2020). Pathogenicity and virulence factors of Fusarium graminearum including factors discovered using next generation sequencing technologies and proteomics. Microorganisms 8:305. doi: 10.3390/microorganisms8020305
Riviezzi, B., García-Laviña, C. X., Morel, M. A., and Castro-Sowinski, S. (2021). Facing the communication between soybean plants and microorganisms (Bradyrhizobium and Delftia) by quantitative shotgun proteomics. Symbiosis 83, 293–304. doi: 10.1007/s13199-021-00758-4
Saima, K., Pandotra, P., Gupta, A. P., Salgotra, R. K., Manzoor, M. M., Lone, S. A., et al. (2017). “Plant molecular breeding: way forward through next-generation sequencing,” in Plant OMICS and Crop Breeding (Apple Academic Press), 201–233.
Sangare, S. S. (2012). Effect of different seed primng methods on germination, seedling establishment and vigour in sorghum Sorghum bicolor (L.) Moench.) and Bambara groundnut (Vigna subterrenea (L.) Verdc.) MSc (Seed Science and Technology), Kwame Nkrumah University of Science and Technology, Kumasi.
Saral, A., Kanekar, S., Koul, K. K., and Bhagyawant, S. S. (2021). Plant growth promoting bacteria induce anti-quorum-sensing substances in chickpea legume seedling bioassay. Physiol. Mol. Biol. Plants 27, 1577–1595. doi: 10.1007/s12298-021-01034-x
Schulte, C. C., Borah, K., Wheatley, R. M., Terpolilli, J. J., Saalbach, G., Crang, N., et al. (2021). Metabolic control of nitrogen fixation in rhizobium-legume symbioses. Sci. Adv. 7:eabh2433. doi: 10.1126/sciadv.abh2433
Shu, H., Luo, Z., Peng, Z., and Wang, J. (2020). The application of CRISPR/Cas9 in hairy roots to explore the functions of AhNFR1 and AhNFR5 genes during peanut nodulation. BMC Plant Biol. 20:417. doi: 10.1186/s12870-020-02614-x
Singh, R. K., Sahu, P. P., Muthamilarasan, M., Dhaka, A., and Prasad, M. (2017). “Genomics-assisted breeding for improving stress tolerance of graminaceous crops to biotic and abiotic stresses: progress and prospects,” in Plant Tolerance to Individual and Concurrent Stresses. ed. Senthil-Kumar, M. (New Delhi: Springer), 59–81.
Singh, B. B., Solanki, R. K., Chaubey, B. K., and Verma, P. (2011). “Breeding for improvement of warm season food legumes,” in Biology and Breeding of Food Legumes. eds. A. Pratap and J. Kumar (Oxfordshire, UK: CABI), 63.
Smalla, K., Sessitsch, A., and Hartmann, A. (2006). The rhizosphere: “soil compartment influenced by the root”. FEMS Microbiol. Ecol. 56:165. doi: 10.1111/j.1574-6941.2006.00148.x
Soumare, A., Diedhiou, A. G., and Kane, A. (2021). Bambara groundnut: a neglected and underutilized climate-resilient crop with great potential to alleviate food insecurity in sub-Saharan Africa. J. Crop Improv. 1–21. doi: 10.1080/15427528.2021.2000908
Staudinger, C., Mehmeti-Tershani, V., Gil-Quintana, E., Gonzalez, E. M., Hofhansl, F., Bachmann, G., et al. (2016). Evidence for a rhizobia-induced drought stress response strategy in Medicago truncatula. J. Proteome 136, 202–213. doi: 10.1016/j.jprot.2016.01.006
Su, P., Zhao, L., Li, W., Zhao, J., Yan, J., Ma, X., et al. (2021). Integrated metabolo-transcriptomics and functional characterization reveals that the wheat auxin receptor TIR1 negatively regulates defense against Fusarium graminearum. J. Integr. Plant Biol. 63, 340–352. doi: 10.1111/jipb.12992
Sugiyama, A. (2019). The soybean rhizosphere: metabolites, microbes, and beyond—A review. J. Adv. Res. 19, 67–73. doi: 10.1016/j.jare.2019.03.005
Swarbreck, S. M., Defoin-Platel, M., Hindle, M., Saqi, M., and Habash, D. Z. (2011). New perspectives on glutamine synthetase in grasses. J. Exp. Bot. 62, 1511–1522. doi: 10.1093/jxb/erq356
Tan, X. L., Azam-Ali, S., Goh, E. V., Mustafa, M., Chai, H. H., Ho, W. K., et al. (2020). Bambara groundnut: an underutilized leguminous crop for global food security and nutrition. Front. Nutr. 7:601496. doi: 10.3389/fnut.2020.601496
Tor-Roca, A., Garcia-Aloy, M., Mattivi, F., Llorach, R., Andres-Lacueva, C., and Urpi-Sarda, M. (2020). Phytochemicals in legumes: a qualitative reviewed analysis. J. Agric. Food Chem. 68, 13486–13496. doi: 10.1021/acs.jafc.0c04387
Trivedi, P., Leach, J. E., Tringe, S. G., Sa, T., and Singh, B. K. (2020). Plant–microbiome interactions: from community assembly to plant health. Nat. Rev. Microbiol. 18, 607–621. doi: 10.1038/s41579-020-0412-1
Turan, M., Kıtır, N., Alkaya, Ü., Günes, A., Tüfenkçi, Ş., Yıldırım, E., et al. (2016). “Making soil more accessible to plants: the case of plant growth promoting Rhizobacteria,” in Plant Growth. Vol. 5. ed. Rigobelo, E. (Croatia: InTechOpen, 61–69.
Udeh, E. L., Nyila, M. A., and Kanu, S. A. (2020). Nutraceutical and antimicrobial potentials of Bambara groundnut (Vigna subterranean): A review. Heliyon 6:e05205. doi: 10.1016/j.heliyon.2020.e05205
Valette, M., Rey, M., Gerin, F., Comte, G., and Wisniewski-Dyé, F. (2020). A common metabolomic signature is observed upon inoculation of rice roots with various rhizobacteria. J. Integr. Plant Biol. 62, 228–246. doi: 10.1111/jipb.12810
Valetti, L., Iriarte, L., and Fabra, A. (2018). Growth promotion of rapeseed (Brassica napus) associated with the inoculation of phosphate solubilizing bacteria. Appl. Soil Ecol. 132, 1–10. doi: 10.1016/j.apsoil.2018.08.017
Valombola, J., Akundabweni, L., Awala, S., and Hove, K. (2019). Agronomic and morphological diversity of Bambara groundnut (Vigna subterranea (L.) Verdc.) accessions in north-Central Namibia. Welwitschia Int. J. Agric. Sci. 1, 88–99.
Verma, S. K., Sahu, P. K., Kumar, K., Pal, G., Gond, S. K., Kharwar, R. N., et al. (2021). Endophyte roles in nutrient acquisition, root system architecture development and oxidative stress tolerance. J. Appl. Microbiol. 131, 2161–2177. doi: 10.1111/jam.15111
Vora, S. M., Ankati, S., Patole, C., Podile, A. R., and Archana, G. (2021). Alterations of primary metabolites in root exudates of intercropped Cajanus cajan–Zea mays modulate the adaptation and proteome of Ensifer (Sinorhizobium) fredii NGR234. Microb. Ecol. doi: 10.1007/s00248-021-01818-4 [Epub ahead of print]
Walker, L., Lagunas, B., and Gifford, M. L. (2020). Determinants of host range specificity in legume-rhizobia Symbiosis. Front. Microbiol. 11:585749. doi: 10.3389/fmicb.2020.585749
Wang, Q., Liu, J., and Zhu, H. (2018). Genetic and molecular mechanisms underlying symbiotic specificity in legume-rhizobium interactions. Front. Plant Sci. 9:1942. doi: 10.3389/fpls.2018.01942
Weinstein, S. B., Martínez-Mota, R., Stapleton, T. E., Klure, D. M., Greenhalgh, R., Orr, T. J., et al. (2021). Microbiome stability and structure is governed by host phylogeny over diet and geography in woodrats (Neotoma spp.). Proc. Natl. Acad. Sci. 118:e2108787118. doi: 10.1073/pnas.2108787118
White, L. J., Ge, X., Brözel, V. S., and Subramanian, S. (2017). Root isoflavonoids and hairy root transformation influence key bacterial taxa in the soybean rhizosphere. Environ. Microbiol. 19, 1391–1406. doi: 10.1111/1462-2920.13602
William, A., George, N., O'reilly Patrick, J., Sean, M., Aryo, F., and Halimi, A. (2016). Adoption of Bambara groundnut production and its effects on farmers welfare in northern Ghana. Afr. J. Agric. Res. 11, 583–594.
Yao, D. N., Kouassi, K. N., Erba, D., Scazzina, F., Pellegrini, N., and Casiraghi, M. C. (2015). Nutritive evaluation of the Bambara groundnut Ci12 landrace [Vigna subterranea (L.) Verdc.] produced in Côte d’Ivoire. Int. J. Mol. Sci. 16, 21428–21441. doi: 10.3390/ijms160921428
Yin, C., Hulbert, S. H., Schroeder, K. L., Mavrodi, O., Mavrodi, D., Dhingra, A., et al. (2013). Role of bacterial communities in the natural suppression of Rhizoctonia solani bare patch disease of wheat (Triticum aestivum L.). Appl. Environ. Microbiol. 79, 7428–7438. doi: 10.1128/AEM.01610-13
Yuan, Y., Bayer, P. E., Batley, J., and Edwards, D. (2017). Improvements in genomic technologies: application to crop genomics. Trends Biotechnol. 35, 547–558. doi: 10.1016/j.tibtech.2017.02.009
Keywords: Bambara groundnut, beneficial bacteria, food security, microbiome engineering, omics
Citation: Ajilogba CF, Olanrewaju OS and Babalola OO (2022) Improving Bambara Groundnut Production: Insight Into the Role of Omics and Beneficial Bacteria. Front. Plant Sci. 13:836133. doi: 10.3389/fpls.2022.836133
Edited by:
Mariela I. Monteoliva, Instituto Nacional de Tecnología Agropecuaria (INTA), ArgentinaReviewed by:
Anukool Vaishnav, Agroscope, SwitzerlandVijay S. Meena, The International Maize and Wheat Improvement Center (CIMMYT), India
Copyright © 2022 Ajilogba, Olanrewaju and Babalola. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Olubukola Oluranti Babalola, olubukola.babalola@nwu.ac.za
 Caroline Fadeke Ajilogba
Caroline Fadeke Ajilogba Oluwaseyi Samuel Olanrewaju
Oluwaseyi Samuel Olanrewaju Olubukola Oluranti Babalola
Olubukola Oluranti Babalola