The N6-Methylandenosine-Related Gene BIRC5 as a Prognostic Biomarker Correlated With Cell Migration and Immune Cell Infiltrates in Low Grade Glioma
- 1Department of Neurosurgery, the Second Affiliated Hospital of Kunming Medical University, Kunming, China
- 2Key Laboratory of Animal Models and Human Disease Mechanisms of Chinese Academy of Sciences and Yunnan Province, Kunming Institute of Zoology, Kunming, China
- 3Kunming College of Life Science, University of Chinese Academy of Sciences, Beijing, China
- 4Kunming Medical University, Kunming, China
Gliomas account for 75% of all primary malignant brain tumors in adults and are associated with high mortality. Emerging evidence has demonstrated that baculoviral inhibitor of apoptosis repeat containing 5 (BIRC5) plays a critical role in cell apoptosis and the progression of diverse cancers. However, no studies have yet focused on the immunological function and mechanisms of upstream BIRC5 regulation in the progression of low-grade gliomas (LGG). Here, we evaluated BIRC5 expression and clinical characteristics in people with LGG using the Chinese Glioma Genome Atlas, The Cancer Genome Atlas, Gene Expression Omnibus, Rembrandt, and Gravendeel databases. We used Kaplan–Meier statistics and receiver operating characteristic (ROC) curves to analyze the prognostic value of BIRC5 in LGG. Kyoto Encyclopedia of Genes and Genomes (KEGG) and Gene Ontology (GO) enrichment terms were also explored to identify functional roles of BIRC5. The Tumor Immune Estimation Resource (TIMER) and Tumor Immune System Interaction (TISIDB) databases were used to examine the correlation between BIRC5 expression and immune cell infiltration in LGG. The Genomics of Drug Sensitivity in Cancer (GDSC) and Cancer Therapeutics Response Portal (CTRP) databases were used to examine the potential drugs targeting BIRC5. We used transwell and wound healing assays to determine the biological functions of BIRC5 in glioma cell migration. Our results demonstrated that BIRC5 was highly expressed in LGG and the expression level correlated with tumor grade, prognosis, histological subtype, isocitrate dehydrogenase 1 (IDH1) mutation, 1p/19q chromosomal co-deletion, chemotherapy status, and O[6]-methylguanine-DNA methyltransferase (MGMT) promoter methylation status. GO and KEGG analysis showed that BIRC5 is primarily involved in cell proliferation and immune response-related signaling pathways. We also found that BIRC5 was significantly correlated with m6A modification and diverse drug sensitivity. TIMER and TISIDB database analysis showed that BIRC5 expression is associated with infiltration of diverse immune cells and immune modulation in LGG. BIRC5 knockdown inhibited LGG cell migration. Collectively, our results demonstrate that BIRC5 is correlated with cell migration and immune infiltration in LGG and may be a useful prognostic biomarker.
1 Introduction
Low-Grade Glioma (LGG) is a relatively common tumor in the central nervous system that mainly includes World Health Organization (WHO) grade 2 and 3 gliomas (Ostrom et al., 2014). Mounting evidence has demonstrated that the molecular characteristics of gliomas include mutated isocitrate dehydrogenase 1 and 2 genes (IDH1/2) and co-deletion of 1p/19q (Aldape et al., 2019). Despite surgical advances and progress in adjuvant therapeutic strategies, clinical outcomes in people with LGG are still poor. Therefore, uncovering the molecular mechanisms underlying the initiation and progression of LGG and identifying highly reliable biomarkers is crucial to improving the diagnosis and treatment of people with this cancer type.
Members of the BIRC gene family (BIRC1–8) mainly encode inhibitors of apoptosis (LaCasse et al., 1998; Makuch-Kocka et al., 2021). These genes are reported to play important roles in cell cycle regulation, immune system activities, and signal transduction (Liang et al., 2020). Aberrant expression of these genes also contributes to the progression of diverse cancers: for instance, OCT4 elevates the promoter activity of BIRC5, contributing to the progression of hepatocellular carcinoma (Cao et al., 2013). It has been shown that BIRC5 is highly expressed in breast cancer, and its expression is correlated with relapse-free survival and overall survival; it, therefore, may have potential as a useful predictive marker and therapeutic target in this context (Dai et al., 2020). Furthermore, a recent study indicated that microRNA-203 inhibits the expression of BIRC5 and suppresses the proliferation and migration of human triple-negative breast cancer cells (Wang et al., 2012). Multiple studies have found that BIRC genes have a pivotal role in multiple physiological processes including cell growth, cell cycle regulation, immune responses, and cancer progression (Pu et al., 2020). However, no studies have yet focused on the immunological function and upstream regulatory mechanism of BIRC genes in the progression of LGG: in this study, we therefore aimed to investigate the role of BIRC genes in these processes.
By integrated analysis of the genomic, transcriptomic, and clinical data from the Chinese Glioma Genome Atlas (CGGA), The Cancer Genome Atlas (TCGA), Gene Expression Omnibus (GEO), Rembrandt and Gravendeel datasets, we found that the RNA and protein of BIRC5 were significantly up-regulation in LGG tissue, high level of BIRC5 was correlated with tumor grade and poor prognosis in LGG. Additionally, BIRC5 expression is also associated with histology subtypes, IDH1 mutation, 1p/19q chromosomal co-deletion, chemotherapy status, and MGMT promoter methylation status, well-established molecular characteristics of gliomas. The potential signaling pathway participated by BIRC5 in LGG was examined by GSEA. Furthermore, our results demonstrated that BIRC5 expression in LGG was regulated by DNA hypo-methylation, the correlation between BIRC5 and immune cell infiltration was evaluated. Finally, qRT-PCR was used to validate the expression of BIRC5 in normal human astrocytes cells and glioma cell lines. Knock-down of BIRC5 significantly inhibits the cell migration of gliomas cells in vitro. The findings in our study demonstrated that the crucial role of BIRC5 in LGG patients, and discovered an underlying mechanism between BIRC5 and immune response.
2 Materials and Methods
2.1 Gene Expression and Survival Analysis
We download the RNA expression and clinical data of glioma cohort projects from The Cancer Genome Atlas (TCGA) research program (https://www.cancer.gov/tcga), including 505 cases of lower-grade glioma (LGG) and 155 cases of GBM RNA sequencing counts data with clinical information. The various normal tissues expression data obtained from the Genotype-Tissue Expression Project (GTEx) RNA sequencing data as resources (https://www.genome.gov/Funded-Programs-Projects/Genotype-Tissue-Expression-Project). This dataset was utilized analysis the expression of BIRC5 in glioma and used to examine the correlation between BIRC5 and various clinical features. We also download the glioma RNA-seq dataset “mRNAseq_693” recruited in the Chinese Glioma Genome Atlas (CGGA) (HTTP: //www.cgga.org.cn/analyse/RNA-data.) contains 693 glioma samples were utilized as verification of the expression and prognosis of BIRC5 in glioma. The GSE4290, GSE16011 andGSE50161 dataset was downloaded from The Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/) database, used to analyze the expression of BIRC5 in low-grade glioma.
2.2 GEPIA Analysis
The GEPIA website (http://gepia.cancer-pku.cn/) was used to explore prognosis of BIRC family gene in LGG. Furthermore, we used GEPIA database examine the correlation between BIRC5 expression and pathological stage of various human cancers.
2.3 Univariate and Multivariate Cox Analyses
Univariate and multivariate Cox analyses were conducted by using the R package “survival.” The WHO grade, IDH status, 1p/19q codeletion, primary therapy outcome, gender, age, and BIRC5 expression were included in these analyses. The receiver operating characteristic (ROC) analysis was performed with the R package “pROC.”
2.4 Gene Set Enrichment Analysis and Kyoto Encyclopedia of Genes and Genome Analyses
Gene expression data of LGG in HTSeq-Counts were downloaded from TCGA website for further analysis. The correlated genes of BIRC5 were screened with Pearson’s correlation coefficients (|r| >0.6 and p < 0.001) with R package “deseq2.” According to the default statistical methods, an adjusted p-value < 0.05 was considered significant. GO and KEGG analysis were conducted on BIRC5 correlated genes with R package “clusterProfiler” to identify the possible biological functions and signaling pathways affected by BIRC5. In the present research, the gene set kegg.v6.2.symbols.gmt, which served as a reference gene set, was downloaded from the Molecular Signatures Database (MSigDB) (http://software.broadinstitute.org/gsea/msigdb). We performed GSEA to examine the potential signaling pathway participated by BIRC5 in low-grade glioma.
2.5 Tumor Immune Estimation Resource
TIDIDB (http://cis.hku.hk/TISIDB/), an integrated repository portal for tumor-immune system interactions, utilized to analysis of the expression of BIRC5 in diverse immune subtype of low-grade glioma (Ru et al., 2019). TIMER (https://cistrome.shinyapps.io/timer/) (Li et al., 2017), an interactive web portal, could perform a comprehensive analysis on the infiltration levels of different immune cells. In this study, we used the TIMER database to explore the correlation between BIRC5 and diverse immune cell infiltration, and different immune-related gene marker in low-grdae glioma.
2.6 DNA Methylation Analysis for BIRC5
We utilized MethSurv (https://biit.cs.ut.ee/methsurv/), a web tool to perform multivariable survival analysis using DNA methylation data, to analyze the diverse DNA methylation sites in the promoter of BIRC5 in glioma (Modhukur et al., 2018). Furthermore, the SMART (http://www.bioinfo-zs.com/smartapp/), an interactive web application for comprehensive DNA methylation analysis and visualization, was used to examine the correlation between BIRC5 expression and DNA methylation in glioma (Li et al., 2019).
2.7 Analysis of the Correlation Between the BIRC5 Expression and Drug Sensitivity
We utilized the GDSC (https://www.cancerrxgene.org/) and CTRP (http://portals.broadinstitute.org/ctrp.v2.1/) databases to analyze the correlation between BIRC5 expression and drug sensitivity (Basu et al., 2013; Yang et al., 2013).
2.8 Cell Culture, RNA Isolation, and Real-Time PCR
The gliomas cell lines were purchased from the cell bank of Kunming Institute of Zoology, and cultured in DMEM medium (Corning) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin. HEK-293T cells were cultured in DMEM medium (Corning). Total RNA was extracted according to the manufacturer’s protocol, and then reverse-transcribed using RT reagent Kit (Takara Bio, Beijing, China, Cat# RR047A; TIANGEN Biotech, Beijing, China, Cat# KR211-02). Real-time PCR was performed by FastStart Universal SYBR Green Master Mix (Roche, Cat# 04194194001; TIANGEN Biotech, Beijing, China, Cat# FP411-02) using an Applied Biosystems 7500 machine. The primers used in this study are shown in the following, BIRC5-F: AGGACCACCGCATCTCTACAT, BIRC5-R: AAGTCTGGCTCGTTCTCAGTG; β-actin-F: CTTCGCGGGCGACGAT, β-actin-R: CCATAGGAATCCTTCTGACC. The expression quantification was obtained with the 2−ΔΔCt method.
2.9 Plasmid Construction and siRNA Interference
The BIRC5 siRNA, control siRNA were synthesized by GenePharma (Shanghai, China), and a scrambled siRNA was synthesized as a negative control. Transfection was performed using Lipofectamine 3000 (Invitrogen, Carlsbad, CA, United States) according to the manufacturer’s instructions. Total RNA was collected 48 h after transfection. All the sequences used in the present study are the following: BIRC5 siRNA: GAAACTGCGGAGAAAGTGCGC. Scrambled siRNAs were used as negative control.
2.10 Cell Migration Assays
Cell migration assay was performed as previously described (Xiong et al., 2021). In this study, GBM cells, including U251 and A172 cells were utilized to determine the function of BIRC5 on the LGG cell migration. Briefly, indicated cells were seeded into 6-well plates (2 × 106/cell) and incubated for 1 day, and then a straight line was scraped with pipette tips. Detached cells were removed. Photographs were taken at the indicated time, and the relative traveled distance was measured. For the trans-well migration assay, 2 × 104 cells/well in 100 μl serum-free medium were plated in a 24-well plate chamber insert, and the lower chamber was filled with 10 % FBS. After incubation for 24 h, cells were fixed with 4% PFA, washed, and then stained with 0.5% crystal violet for further pictures captured.
2.11 Immunohistochemistry Assay
Immunohistochemistry was performed as described previously (Xu et al., 2020). Briefly, paraffin sections were deparaffinized by xylene, rehydrated with gradient ethanol, and subjected to antigen retrieval. After H2O2 treatment and blockage with 10% normal goat serum, the slides were incubated with indicated antibodies (Proteintech, BIRC5 Polyclonal Antibody, Rabbit Polyclonal,Catalog number: 10508-1-AP, 1:200) followed by an incubation with biotinylated secondary antibody and streptavidin-HRP (Dako, K5007).
2.12 Statistical Analysis
The survival data from the CGGA database were acquired by KM. Correlation analysis was performed using the Pearson correlation test. Kaplan-Meier survival curves were plotted to exhibit the overall survival for low-grade glioma patients. Univariate and multivariate Cox regression analyses were used to examine the independent prognostic significance of each variable enrolled in this finding. The significance of the data between the two experimental groups was determined by Student’s t-test, and multiple group comparisons were analyzed by one-way ANOVA. p < 0.05 (*), p < 0.01 (**) and p < 0.001 (***), were considered significant.
3 Results
3.1 Expression and Prognostic Value of BIRC Family Genes in Low-Grade Gliomas
To explore the functions of the BIRC gene family in LGG, we first examined the expression of BIRC1–8 using the TCGA and GTEx datasets. BIRC1-BIRC7 was found to be significantly up-regulated in LGG; BIRC8 genes showed stable expression levels, with no significant difference in expression between LGG and controls (Supplementary Figure S1A). We also used the GEPIA database to determine the prognostic value of BIRC family genes in LGG; among these genes, increased BIRC5 expression was correlated with worse clinical outcomes in this cancer type (Supplementary Figure S1B). Overall, these results indicated that BIRC5 was highly expressed in LGG with high specificity.
3.2 BIRC5 is Highly Expressed in Human Cancer and is Related to Patient Prognosis
We used the TGCA database to examine the expression of BIRC5 in all cancers. Our results demonstrated that BIRC5 was significantly overexpressed in BLCA, BRCA, CHOL, COAD, ESCA, HNSC, KICH, KIRC, KIRP, LIHC, LUAD, LUSC, PRAD, READ, STAD, THCA, and UCEC (Figure 1A). Given that BIRC5 was found to be highly expressed in diverse cancer types, we used the kmplot database to examine the prognostic value of BIRC5 across human cancers. The analysis revealed that high levels of BIRC5 expression closely correlated with worse clinical prognosis in ACC, KIRC, KIRP, LGG, LIHC, LUAD, and MESO. In contrast, high levels of BIRC5 expression were associated with improved prognosis in OV (Figure 1B). Together, these findings indicate that BIRC5 expression is closely associated with prognosis in diverse human cancers.
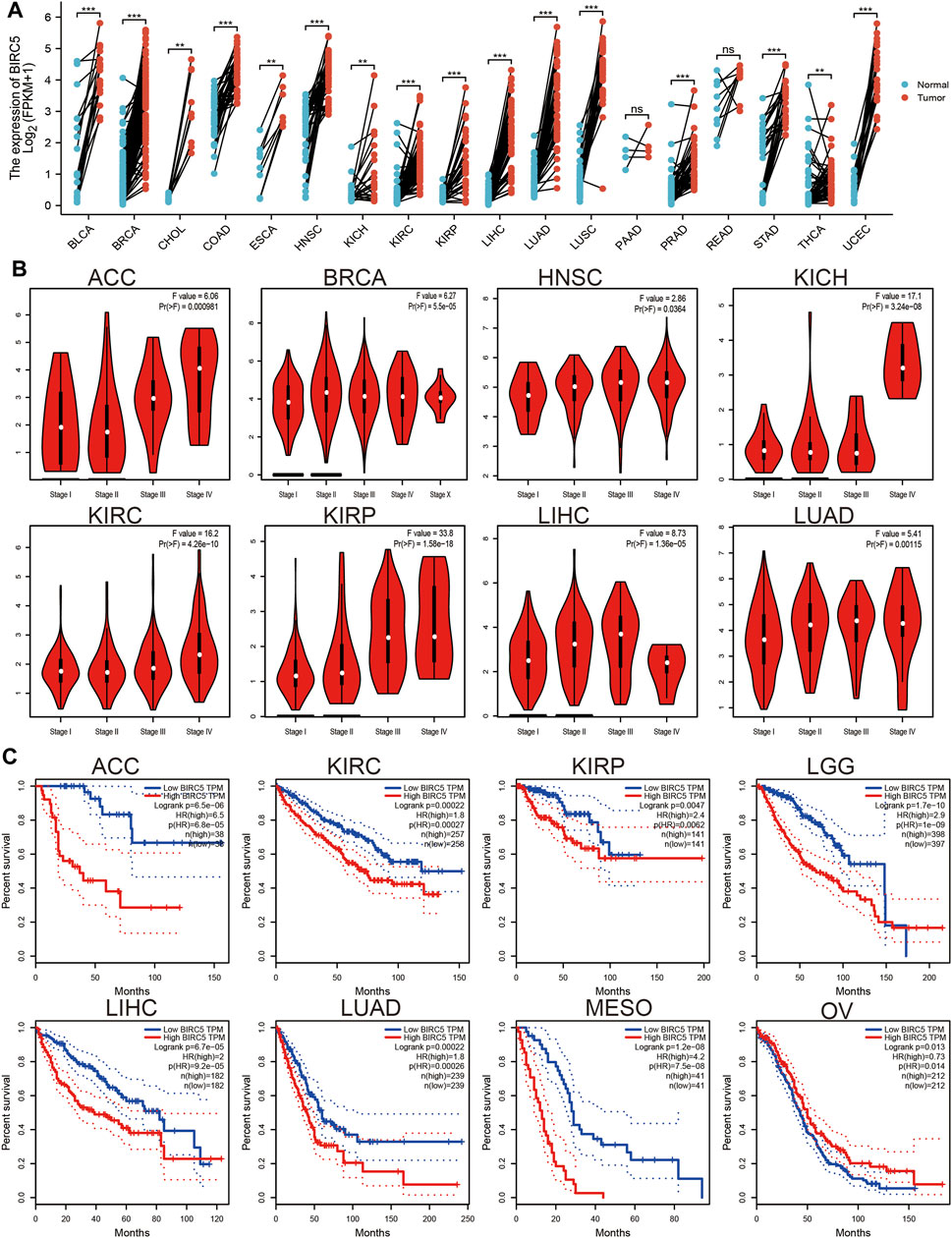
FIGURE 1. BIRC5 expression and prognosis in patients with other cancer types. (A) Differences in expression of BIRC5in normal and tumor tissues of TCGA. (B) The pathological stage of BIRC5 in diverse cancer examine by UALCAN database. (C) Kaplan-Meier curves were drawn to evaluate the overall survival of BIRC5 in various human cancers.
3.3 BIRC5 is Highly Expressed in Glioma and is Related to Patient Prognosis
To examine the expression of BIRC5 in LGG tissue compared with a healthy control group, we used the public bioinformatics databases, including TCGA, GEO, Rembrandt, and Gravendeel. The results showed that BIRC5 was significantly elevated in LGG tissue compared with controls (Figures 2A,B).
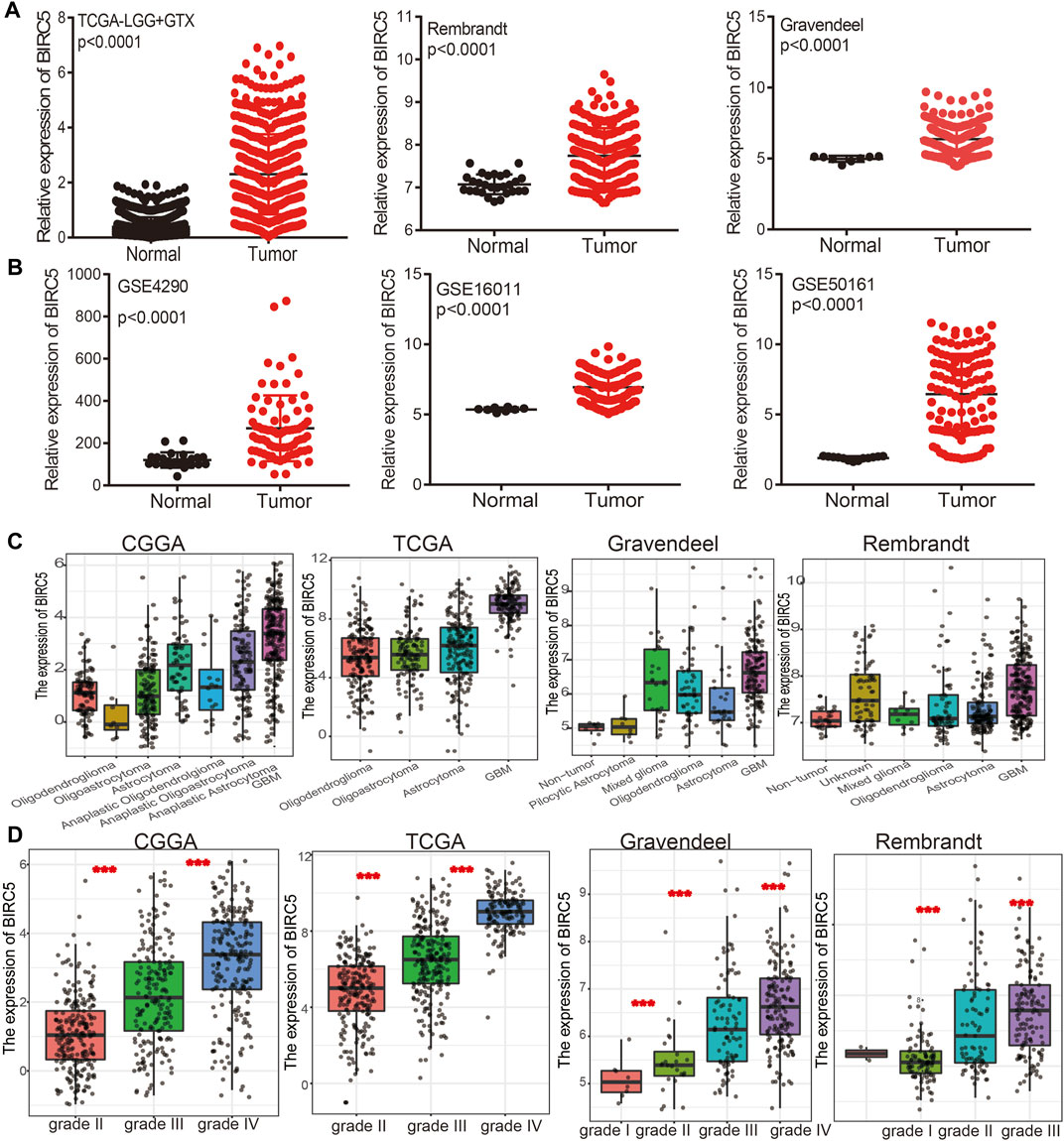
FIGURE 2. BIRC5 was highly expressed in low grade glioma. (A,B) BIRC5 was upregulated in glioma examined by the TCGA, Rembrandt, Gravendeel, and GEO datasets. (C,D) BIRC5 expression levels among glioma patients with different histological subtype (C) and tumor grade (D) in TCGA-LGG.
We further analyzed the correlation between BIRC5 expression levels and clinical characteristics of LGG, focusing primarily on tumor grade, histological subtype, chemotherapy status, IDH1 mutation, 1p/19q chromosomal co-deletion, and O[6]-methylguanine-DNA methyltransferase (MGMT) promoter methylation. Our results showed that BIRC5 was differentially expressed in different histological subtypes of LGG (Figure 2C). BIRC5 expression levels increased at higher tumor grades (Figure 2D). Interestingly, we found that BIRC5 was markedly decreased in the group with IDH mutations and the group with 1p/19q chromosome co-deletion (Figure 3A and Table 1). Additionally, BIRC5 expression was markedly increased in those receiving chemotherapy and those with unmethylated MGMT promoters. These results indicate that BIRC5 plays a major role in LGG development.

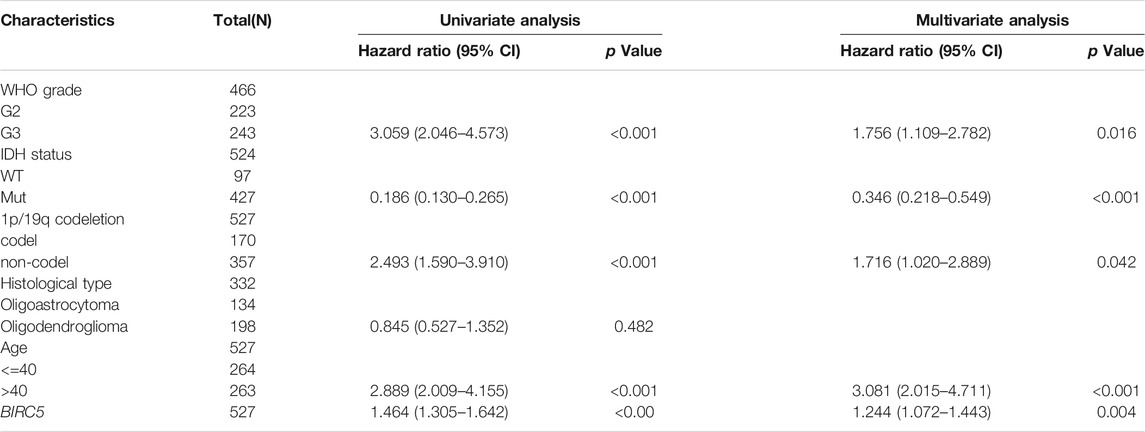
FIGURE 3. Correlation between the BIRC5 expression and clinical information in LGG. (A) The expression of BIRC5 in diverse clinical information of glioma based on CGGA databases. (B) The overall survival between the high- BIRC5and low- BIRC5 in TCGA, CGGA, Rembrandt, and Gravendeel databases. (C) Diagnostic efficacy of the ROC curve of BIRC5 expression in TCGA-LGG, CGGA, and TCGA-GBM.
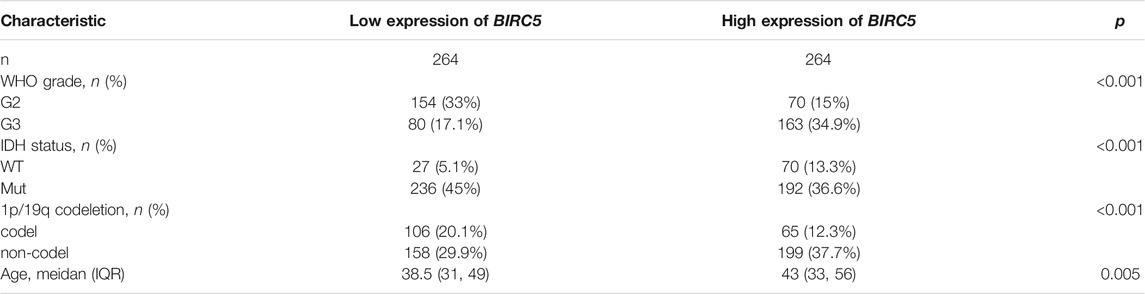
TABLE 1. Correlation between BIRC5 expression and clinicopathological factors based on the TCGA database.
As showed in the Table 2, patients having integral clinical data were included in further Cox regression analysis. In the Cox univariate regression analysis, we found that high expression of BIRC5, WHO grade, IDH status, 1p/19q codeletion, and age were associated with overall survival in LGG patients. Multivariate Cox analysis further indicated that BIRC5 (p = 0.004) was an independent prognostic factor for overall survival in LGG patients, along with WHO grade, IDH status, 1p/19q codeletion, and age.
3.4 Prognostic Value of BIRC5 in Low-Grade Gliomas
Given that BIRC5 is highly expressed in LGG, we examined the prognostic value of BIRC5 expression in this cancer type. People with LGG were divided into “high” or “low” expression groups based on the median expression value. High BIRC5 expression was significantly associated with unfavorable prognosis in people with LGG in different datasets (Figure 3B). Receiver operator characteristic (ROC) curve analysis showed that the area under the curve (AUC) of BIRC5 was 0.970 in the CGGA dataset, 0.981 in the TCGA-LGG dataset, 0.9933 in the Rembrandt dataset, and 0.998 in the Gravendeel dataset (Figure 3C). These results indicate that BIRC5 is a highly sensitive and specific marker with the potential to be used in LGG diagnosis.
3.5 Correlation Between DNA Methylation and BIRC5 Expression in Low-Grade Gliomas
DNA methylation is crucial for the epigenetic regulation of gene expression. To elucidate the mechanisms of abnormal upregulation of BIRC5 in LGG tissues, we explored the correlation between DNA methylation and BIRC5 expression in LGG using diverse public databases. Firstly, we found that BIRC5 methylation was lower in LGG than in healthy brain tissue and was negatively correlated with tumor grade in LGG (Figures 4A,B). Treatment with 5-azacytidine, an inhibitor of DNA methyltransferase (Xiong et al., 2021), resulted in increased BIRC5 RNA levels in U251 human brain cells (Figure 4C). Moreover, we found that cg24107848 methylation was significantly negatively correlated with the BIRC5 expression in LGG (r = −0.340, p < 0.001) (Figure 4D). We also showed that decreased methylation levels at cg28734000 correlated with worse prognosis in the CGGA-LGG dataset using the MethSurv statistical tool (Figures 4E,F). These results suggested that hypomethylation in the BIRC5 promoter region results in increased expression of this gene in LGG.
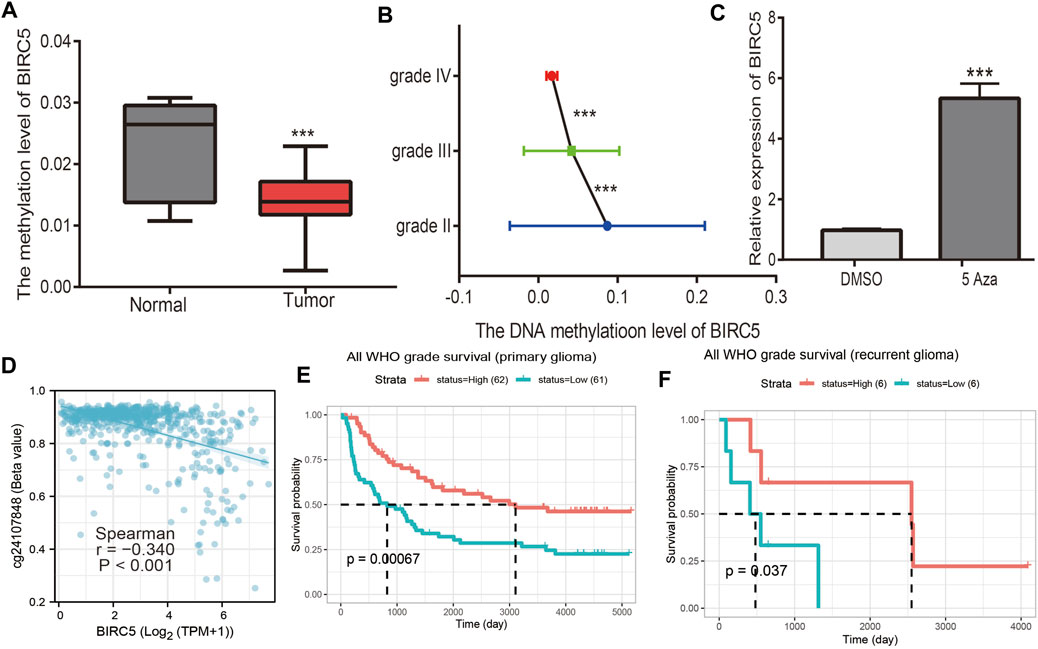
FIGURE 4. Analysis of the BIRC5 methylation level. (A) The DNA methylation level of BIRC5 in LGG. (B) The DNA methylation level of BIRC5 in diverse tumor grades of LGG based on the CGGA dataset. (C) The expression of BIRC5 in U251 cells after using 5-AZA treatment was examined by qRT-PCR assay. (D) The correlation between DNA methylation and BIRC5 expression in low grade glioma by Pearson’s correlation. (E,F) The prognosis for methylation level of BIRC5 in glioma based on the CGGA dataset.
3.6 BIRC5 Expression is Correlates with Regulators of N6-methyladenosine (m6A) RNA Methylation in Low-Grade Gliomas
Given that N6-methyladenosine (m6A) RNA methylation plays a major role in the initiation and progression of LGG, we examined the correlation between BIRC5 expression and regulators of m6A RNA methylation in this cancer type (Figure 5A). BIRC5 expression was significantly positively correlated with 16 m6A-related genes, including HNRNPA2B1 (r = 0.52, p = 0), HNRNPC (r = 0.0.32, p < 0.001), IGF2BP2 (r = 0.32, p <0.001), IGF2BP3 (r = 0.39, p = 0), METTL3 (r = 0.20, p < 0.001), RBM15 (r = 0.46, p = 0), RBM15B (r = 0.48, p = 0), RBMX (r = 0.41, p = 0), WTAP (r = 0.25, p < 0.001), YTHDC2 (r = 0.26, p < 0.001), ALKBH5 (r = 0.21, p = 0.001), YTHDF1 (r = 0.25, p = 0.001), YTHDF2 (r = 0.47, p = 0) and YTHDF3 (r = 0.24, p = 0.001) (Figures 5B–E). Collectively, these results demonstrate that BIRC5 plays indispensable roles in LGG progression via interaction with regulators of m6A RNA methylation.
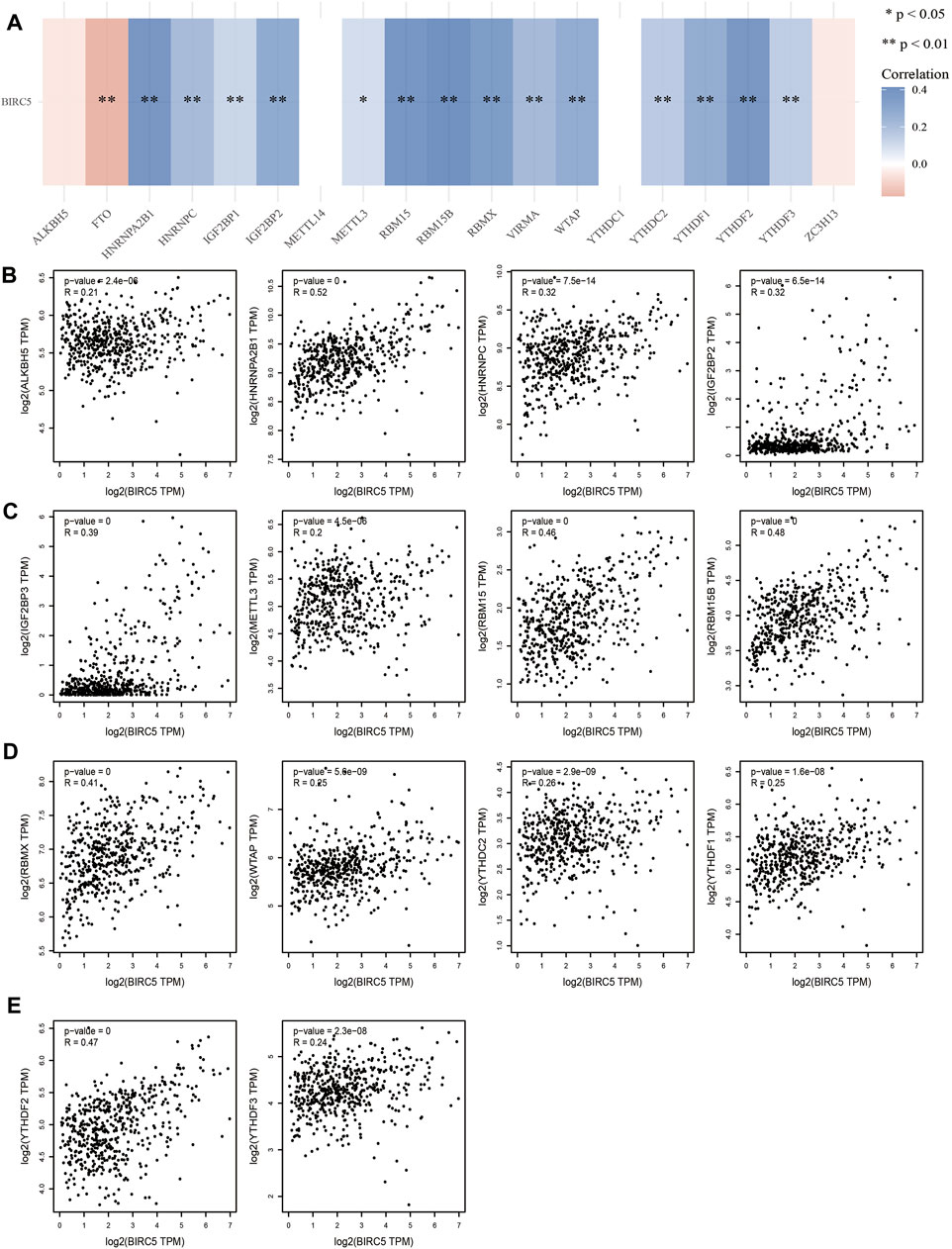
FIGURE 5. Analysis of the correlation between BIRC5 expression and m6A regulator in LGG. (A–E) Analysis of the correlation between BIRC5 expression and m6A regulator in low grade glioma by Pearson’s correlation.
3.7 Functional Analysis of BIRC5 in Low-Grade Gliomas
To explore the specific functions of BIRC5 in LGG progression, we first analyzed genes whose expression was positively correlated with that of BIRC5 in LGG (Figures 6A,B). We then performed Kyoto Encyclopedia of Genes and Genomes (KEGG) and Gene Ontology (GO) enrichment analyses based on these co-expressed genes. The results showed that BIRC5 is primarily involved in tubulin, microtubule, glycosaminoglycan, heparin, and growth factor binding, as well as GABA receptor activity and histone activity in the GO analysis (Figure 6C). BIRC5 may participate in cell cycle, human papillomavirus infection, p53 signaling, extracellular matrix (ECM) receptor interaction, DNA replication, small cell lung cancer, and morphine addiction pathways (Figure 6D). These results demonstrate functionally-relevant roles for BIRC5 in LGG progression.
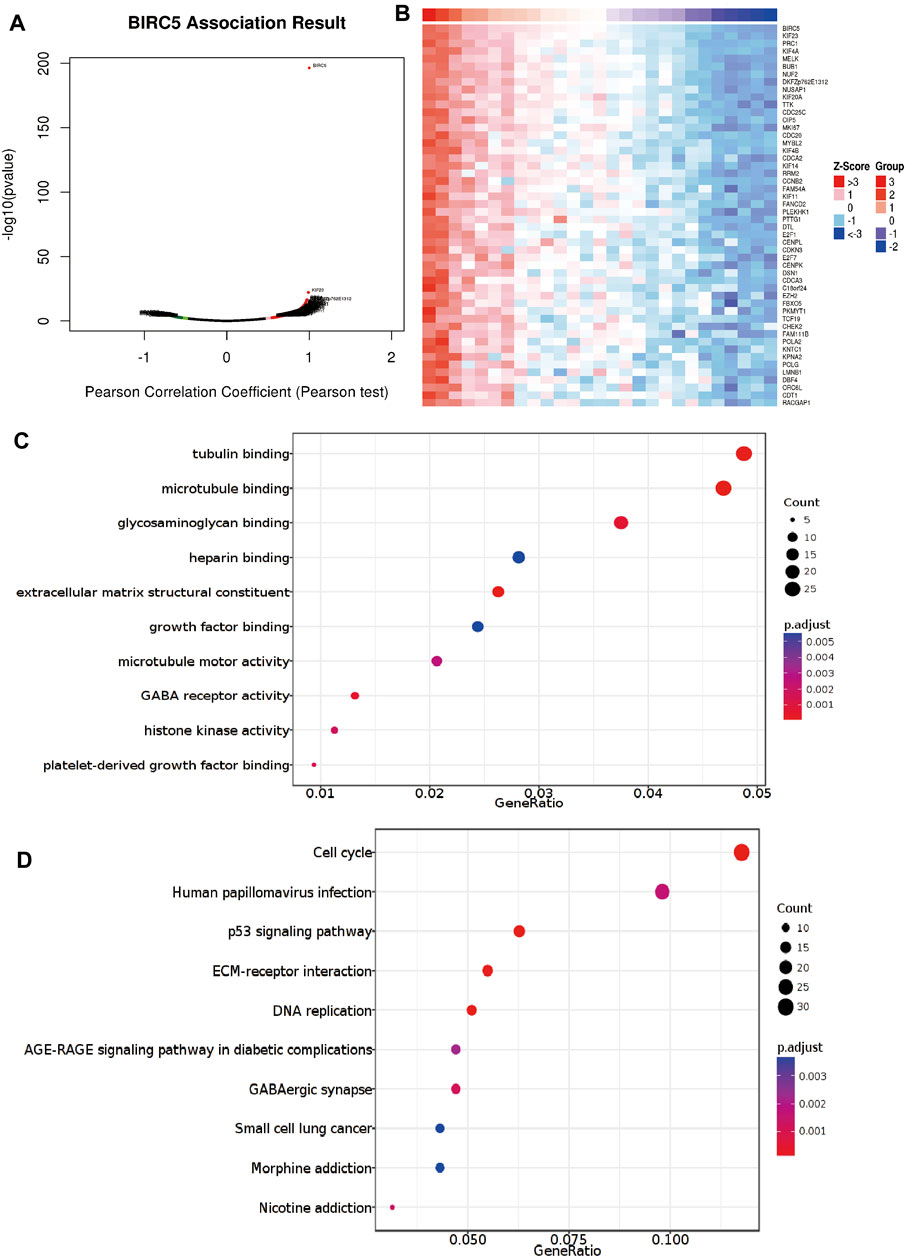
FIGURE 6. Analysis of the function of BIRC5 expression in LGG. (A,B) Analysis of the co-expression gene of BIRC5 in LGG examines by linkomics database. (C) Analysis of the molecular function of BIRC5 in LGG. (D) Analysis of the KEGG signaling pathway of BIRC5 in LGG.
3.8 BIRC5-Related Signaling Pathways Identified Using Gene Set Enrichment Analysis
To identify signaling pathways that are affected by BIRC5 overexpression in glioma, we performed a gene set enrichment analysis (GSEA). High BIRC5 expression was mainly associated with the regulation of stem cell differentiation, cell cycle growth 1/synthesis (G1/S) phase transition, and chromosome segregation (Figure 7A). Moreover, high BIRC5 expression was associated with apoptosis, cell cycle, ubiquitin-mediated proteolysis, and focal adhesion signaling pathways (Figure 7B). Collectively, these results demonstrate that BIRC5 plays an indispensable role in cell proliferation and immune regulation.
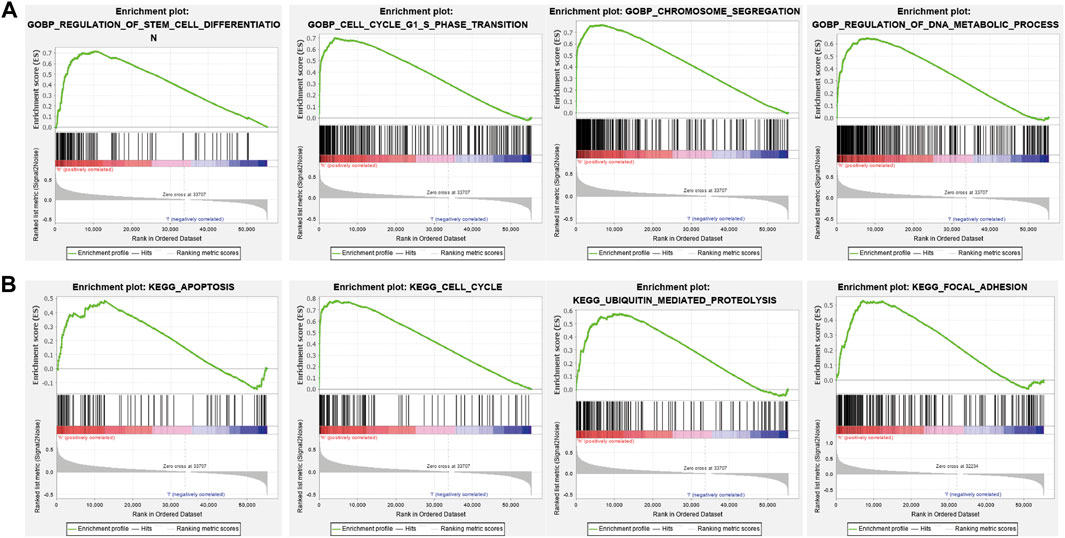
FIGURE 7. The KEGG signal pathway explored by GSEA software. (A) The biological process involved by BIRC5 in LGG was examined by GSEA software. (B) The signaling pathway involved by BIRC5 in LGG was examined by GSEA software.
3.9 Association Between BIRC5 Expression Levels and Immune Cell Infiltration in Low-Grade Gliomas
The level of immune infiltration affects clinical outcomes from tumors. We, therefore, analyzed the expression of BIRC5 in diverse immune subtypes, which showed that BRIC5 expression was highest in the C4 subtype (Figure 8A). Furthermore, we found that different somatic BIRC5 copy number alterations affected infiltration levels of diverse immune cells, including B cells, CD4+ and CD8+ T cells, neutrophils, and dendritic cells (Figure 8B). Finally, we using the TIMER database to examine the correlation between BIRC5 expression and immune infiltration levels for diverse immune cells in LGG; results indicated that BIRC5 was significantly correlated with the level of B cells (cor = 0.271 p < 0.001), CD8+ T cells (cor = 0.158, p < 0.001), CD4+ T cells (cor = 0.208, p < 0.001), macrophages (cor = 0.224, p < 0.001), neutrophils (cor = 0.207, p < 0.001), and dendritic cells (cor = 0.279, p < 0.001), and positively correlated with tumor purity (cor = 0.165, p < 0.001) (Figure 8C). Our Cox proportional hazard model analysis demonstrated that B cells, CD8+ and CD4+ T cells, macrophages, neutrophils, dendritic cells, and BIRC5 expression were significantly associated with worse overall survival in LGG patients (Figure 8D).
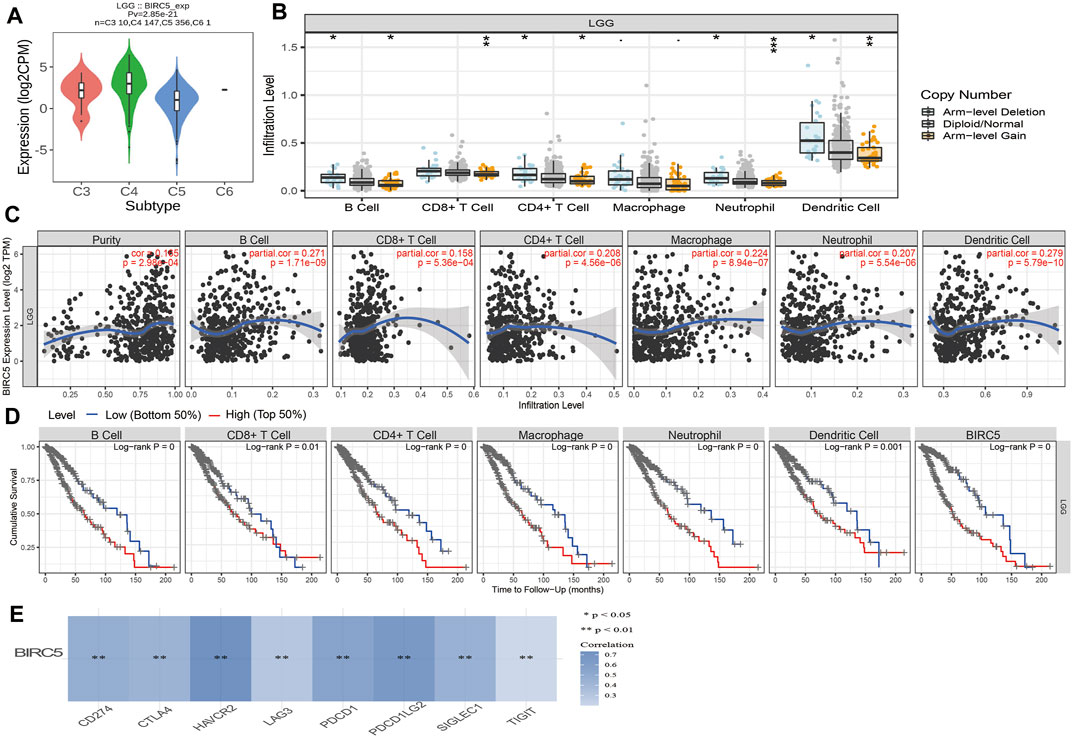
FIGURE 8. Analysis of the correlation between BIRC5 expression and diverse immune cell infiltration. (A) The expression of BIRC5 in diverse immune subtypes of LGG was examined by TISIDB. (B) The correlation between BIRC5 expression and somatic copy number alterations examine by TIMER database. (C) The correlation between BIRC5 expression and diverse immune cell infiltration was examined by TIMER database. (D) The B cells, CD4+ T cells, CD8+ T cells, dendritic cells, Macrophages, and Neutrophils are correlated with the cumulative survival rate in LGG examine by TIMER database. (E) The correlations between BIRC5 expression and diverse immune checkpoints related gene examined by TIMER database.
Given that immune checkpoints play a crucial role in tumor immunosuppression, we analyzed the correlation between BIRC5 expression and that of the immune checkpoint-related genes CD274, CTLA4, HAVCR2, LAG3, PDCD1, PDCD1LG2, TIGIT, and SIGLEC15 in LGG using Pearson correlation analysis. BIRC5 expression was significantly positively correlated with the expression of all eight genes in this analysis (Figure 8E), supporting a role for BIRC5 in immune regulation.
3.10 Correlation Between BIRC5 Expression and Drug Sensitivity
Given that BIRC5 has potentially promoted LGG progression, it is therapeutically useful to identify anti-LGG drugs targeting BIRC5. Therefore, we used GSCA tools to analyze the relationship between BIRC5 expression and sensitivity to anti-LGG drugs. BIRC5 expression was positively correlated with drug sensitivity for trametinib, selumetinib, RDEA119, AZ628, PD-0325901, and VX-11e (all r > 0.14, p < 0.001), and negatively associated with drug sensitivity for NSC-207895, navitoclax, vorinostat, NPK76-II-72-1, and KIN001-270 (all r < −0.10, p < 0.001) in the Genomics of Drug Sensitivity to Cancer (GDSC) database (Figure 9A and Table 3). Similarly, CTRP database analysis demonstrated that BIRC5 expression was positively correlated with drug sensitivity for trametinib, selumetinib, PD318088, MLN2480, BYL-719, PD 153035 and GANT-61 (all r > 0.10, p < 0.001), and negatively associated with the drug sensitivity of BRD-K30748066, docetaxel, GSK-J4, niclosamide, STF-31, GSK461364, SID 26681509, tivantinib, BI-2536, ceramic-2, neopeltolide, CAY10618, fluvastatin, 3-Cl-AHPC and methotrexate (r < −0.14, p < 0.001) (Figure 9B and Table 4). Taken together, the results demonstrate that BIRC5 was correlated with sensitivity to diverse drugs from the Cancer Therapeutic Response Portal database.
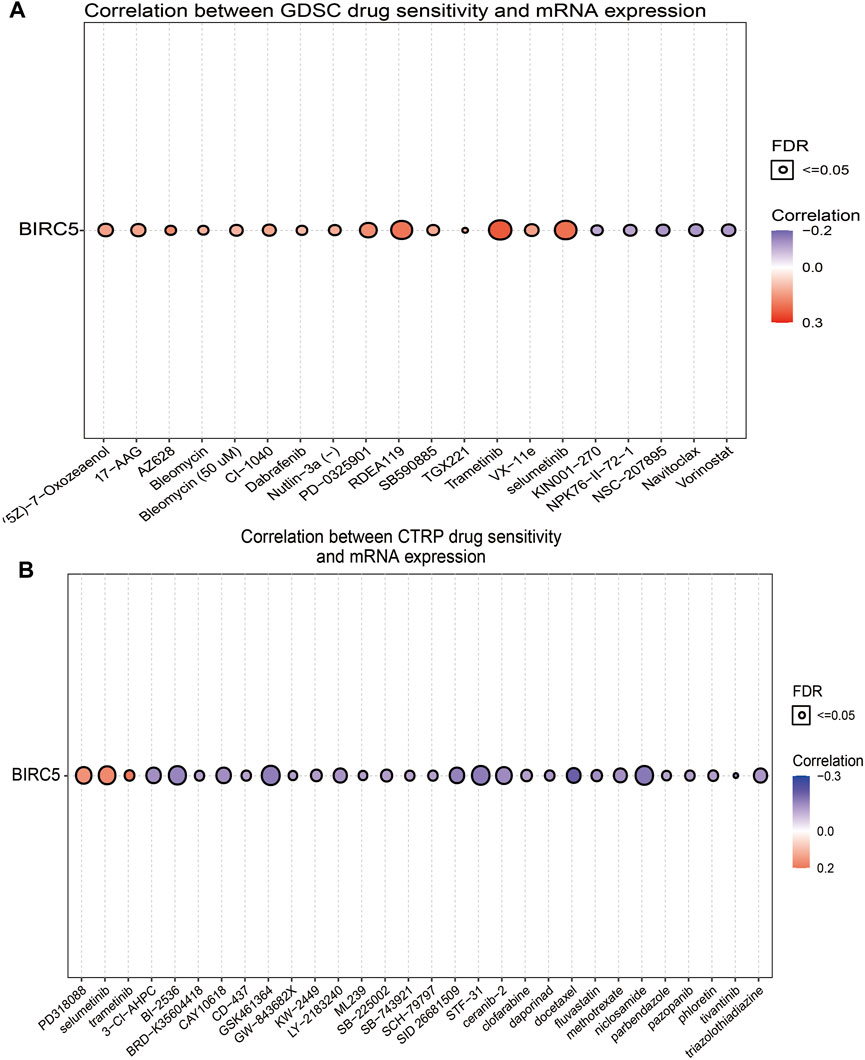
FIGURE 9. Correlation between drug sensitivity and BIRC5 expression. (A,B) The correlation between BIRC5 expression and drug sensitivity in pan-RCC was studied by Spearman correlation analysis. Red represents positive correlation between drugs sensitivity and BIRC5 expression, and blue represents negative correlation between drugs sensitivity and BIRC5 expression. (A): GDSC, (B): CTRP.
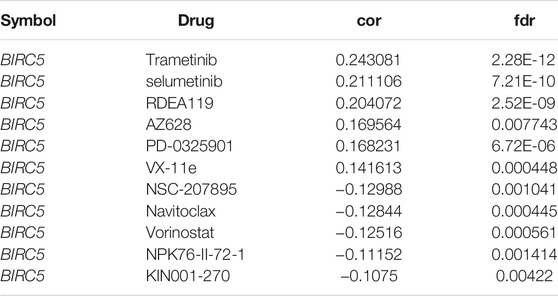
TABLE 3. Analysis of the correlation between BIRC5 expression and diverse drug sensitivity by using GDSC database.
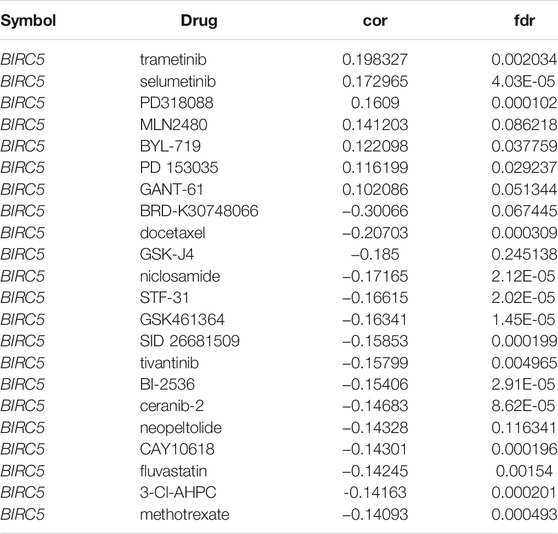
TABLE 4. Analysis of the correlation between BIRC5 expression and diverse drug sensitivity by CTRP database.
3.11 Knockdown of BIRC5 Inhibits Glioma Cell Migration In Vitro
Our KEGG enrichment results showed that BIRC5 may be involved in the focal adhesion signaling pathway, therefore, we used a functional assay to verify this result. We used a quantitative real-time polymerase chain reaction (qRT-PCR) and Western blot assay to detect BIRC5 expression in normal human astrocyte cells and glioma cell lines. BIRC5 was highly expressed in glioma cell lines, especially in U251 cells (Figures 10A,B). IHC assay results also indicated that BIRC5 was highly expressed in glioma tissues than paracancerous tissues (Figure 10C). Based on this result, we constructed BIRC5 knockdown cells using qRT-PCR to verify knockdown efficiency (Figures 10D,E). The resultant depletion of BIRC5 was found to significantly inhibit glioma cell migration by transwell and wound healing assays (Figures 10F–I). Collectively, these results demonstrate that BIRC5 is highly expressed in glioma cells, likely resulting in increased cell migration.
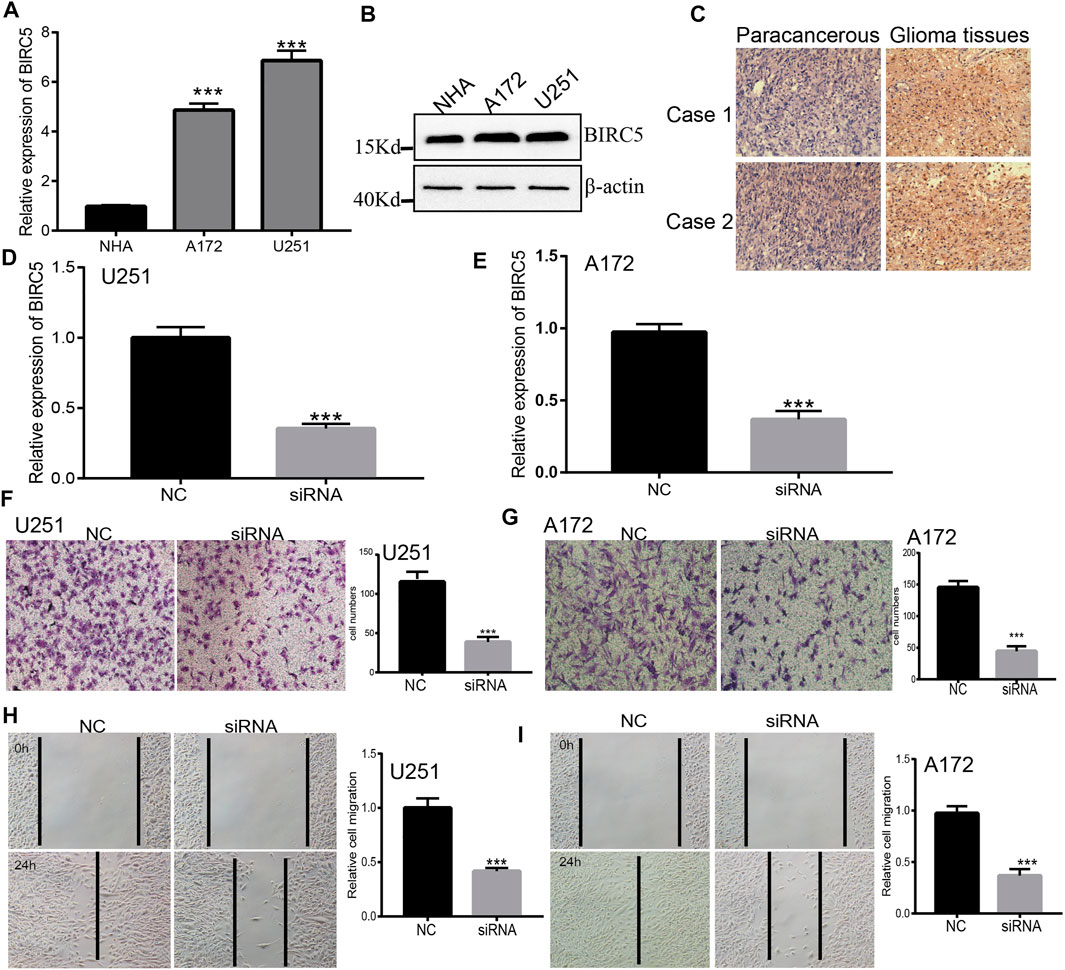
FIGURE 10. Depletion of BIRC5 inhibits glioma cells migration. (A,B) The expression of BIRC5 in normal human astrocytes cells (NHA) and glioma cell lines (A172 and U251) examined by qRT-PCR and Western blot assay. (C) IHC analysis of BIRC5 expression in glioma cancer tissues. Scale bar, 50 μm. (D,E) The expression of BIRC5 in U251 and A172 cells treated with siRNAs (siRNA -BIRC5#NC, siRNA-BIRC5#) measured by qRT–PCR. *p < 0.05, one-way ANOVA test for the comparison of the two groups. (F–I) Transwell and Wound healing assay of the glioma cancer cell lines U251 and A172 treated with shRNAs. *p < 0.05, one-way ANOVA test for the comparison of the two groups. Scale bar, 100 μm.
4 Discussion
Glioma is a highly fatal primary tumor of the central nervous system. Despite diverse available therapeutic strategies, including surgery, chemotherapy, and immunotherapy, the median survival of people with glioma—which has barely improved since its recognition as a separate cancer type—remains only approximately 14 months (Huang et al., 2021). Therefore, more sensitive and specific diagnostic biomarkers and potential therapeutic targets for glioma patients are of no delay.
BIRC5, also called survivin, is an immune-related gene and member of the apoptotic (IAP) protein family (Li et al., 1998). BIRC5 has been shown to play vital roles in carcinogenesis by affect the cell proliferation and by inhibits cell apoptosis (Yamamoto et al., 2008). Since BIRC5 is frequently up-regulated in diverse cancers and treatment that targets BIRC5 has been increasingly noticed as a novel strategy for various malignant tumors (Jaiswal et al., 2015). For instance, BIRC5 was increased in triple-negative and BIRC5 knockdown significantly inhibits the proliferation of breast cancer cells, implying that BIRC5 acts like a tumor driver (Wang et al., 2012). Furthermore, BIRC5 expression has been found to confer resistance to chemotherapy and radiation, targeting BIRC5 in experimental models improves survival (Jha et al., 2012). Together, these findings indicate that BIRC5 may not only function as an oncogene, but also as a promising predictive biomarker and potential therapeutic target in cancer. A recent study confirmed that BIRC5 could act as a reliable prognostic indicator in breast cancer patients (Dai et al., 2020), but the expression, clinical significance, and its impact on the immune microenvironment remain largely unclear in low-grade glioma.
Brunicardi et al. found that BIRC5 was highly expressed in pancreatic cancer and maybe as a precision diagnostic biomarker for the diagnosis for pancreatic cancer. In this study, we uncover that BIRC5 expression was upregulated in diverse human cancer and correlated with poor prognosis, especially in glioma (Figure 1). Which further validated by using the GEO and CGGA database (Figure 2), the IHC and Western blotting assay in glioma cell lines and glioma tissues. Furthermore, higher expression of BIRC5 was positively correlates with adverse clinicopathologic features of glioma, including the tumor grade, histological subtype, isocitrate dehydrogenase 1 (IDH1) mutation, 1p/19q chromosomal co-deletion, chemotherapy status, and O[6]-methylguanine-DNA methyltransferase (MGMT) promoter methylation status (Figure 3). Accumulating studies have confirmed that IDH1 mutations are associated with the occurrence and development of glioma, and IDH1 mutations are more common in low grade glioma than in LGG (Miroshnikova et al., 2016). Therefore, we speculated that the higher expression of BIRC5 in low grade glioma patients might due to more low grade glioma patients are IDH wild-type tumors, which expressed higher BIRC5 levels. Moreover, the KM survival curve analyses showed that upregulated BIRC5 expression associated with adverse clinical outcomes in glioma from both TCGA and CGGA datasets. ROC curve analysis showed that the BIRC5 expression level has diagnostic value for gliomas (Figure 3). Similarly, high expression of BIRC5 was associated with overall survival in breast cancer, clear-cell renal cell carcinoma, gastric cancer, and lung cancer (Ni et al., 2019; Wan et al., 2019; Zou et al., 2019; Dai et al., 2020). We also performed a univariate analysis including the prognostic factors for overall survival. We found that high expression of BIRC5 was associated with higher tumor grades, histological type, IDH mutation status, 1p/19q chromosome co-deletion, and primary therapy outcome. Furthermore, we utilized the Cox regression model for multivariate analysis; the results demonstrated that BIRC5 expression, WHO grade, IDH status, 1p/19q codeletion, and age were an independent prognostic factor for overall survival in LGG patients. Similarly, high expression of BIRC5 was an independent prognostic factor for overall survival in clear-cell renal cell carcinoma and endometrial cancer (Chuwa et al., 2016; Wan et al., 2019).
A growing number of studies have reported that DNA methylation plays an important role in gene expression regulation (Yamashita et al., 2018). In this study, we found that the DNA methylation level of BIRC5 was down-regulated in glioma tissues than normal tissues. Furthermore, we showed that the methylation sites (cg24107848) were significantly negatively correlated with the expression of BIRC5 in LGG. More importantly, our results showed that lower methylation levels in cg24107848 were correlated with a worse prognosis in LGG patients. We also used a qRT-PCR assay confirm that hypomethylation in the BIRC5 promoter region resulted in the increased expression of this gene in LGG (Figure 4). Although various molecular mechanisms can lead to increased gene expression, such as the lncRNA/miRNA axis, regulation of transcription factors, and gene copy number amplification, DNA hypomethylation is one of the main regulatory mechanisms of gene expression.
M6A RNA methylation is also known to affect glioma malignancy (Chai et al., 2019). For example, it has been shown that YTHDF2 promotes the CSC liver phenotype and cancer metastasis by modulating the m6A methylation of OCT4 mRNA (Zhang et al., 2020). In hepatocellular carcinoma, YTHDF2 silenced in human HCC cells or ablated in mouse hepatocytes provoked inflammation, vascular reconstruction, and metastatic progression (Hou et al., 2019). A recent finding found that YTHDF2 sequesters m6A-circRNA and is essential for suppression of innate immunity (Chen et al., 2019). In this study, we found that BIRC5 expression is significantly associated with regulators of m6A methylation (Figure 5). Crosstalk between m6A RNA modification and BIRC5 expression in low-grade glioma, therefore, requires further characterization in future studies.
Sun et al. found that BIRC5 was highly expressed in HCC tissues and involved in cell cycle regulation (Wang et al., 2011). To explore the possible mechanism of BIRC5 affecting the survival of patients, KEGG and GSEA enrichment analyses were performed based on the differentially expressed genes between BIRC5 groups, and the results suggested that higher BIRC5 expression was mainly associated with the regulation of stem cell differentiation, cell cycle growth 1/synthesis (G1/S) phase transition, and chromosome segregation (Figure 6), which is consistent with previous studies that BIRC5 regulated the stem cell differentiation (Lin et al., 2020). Moreover, high BIRC5 expression was associated with apoptosis, cell cycle, ubiquitin-mediated proteolysis, and focal adhesion signaling pathways. Collectively, these results demonstrate that BIRC5 plays an indispensable role in cell proliferation and immune regulation (Figure 7).
Mounting evidence demonstrates that the tumor microenvironment has a key role in the development of tumors, and can be used as a biomarker for diagnosis and prognosis (Wu and Dai, 2017). CD8 + tumor infiltrating lymphocytes have also been shown to be associated with glioma prognosis (Yu-Ju Wu et al., 2020). In this research, we revealed that BIRC5 somatic copy number alterations affected infiltration levels of various immune cells, including B cells, CD4+ and CD8+ T cells, neutrophils, and dendritic cells. Furthermore, we found that BIRC5 was significantly correlated with the level of B cells, CD8+ T cells, CD4+ T cells, macrophages, neutrophils and dendritic cells, and was negatively correlated with tumor purity. We showed that BIRC5 influences the survival time of LGG patients, partially through immune cell infiltration (Figure 8). These findings indicate that BIRC5 could be a novel immune-related therapeutic target in LGG.
It has been demonstrated that immune modulators play crucial roles in tumor cell evasion of immune surveillance (Xue et al., 2017). Studies have confirmed that the increased of immune checkpoints such as PD-L1, CTLA-4, TIM-3, and LAG3 in glioma helps tumor immune evasion, leading to T cell dysfunction (Xue et al., 2017; Woroniecka et al., 2018). Our results confirmed that BIRC5 expression was significantly positively correlated with the expression of CD274, CTLA4, HAVCR2, LAG3, PDCD1, PDCD1LG2, TIGIT, and SIGLEC15, suggesting a crucial role of BIRC5 in regulating the expression of immune checkpoints and immunotherapy (Figure 8). Given the effects of BIRC5 on LGG immune cell infiltration, we can infer that increased expression of BIRC5 may promote mast cell infiltration and contribute to a poor prognosis. Therefore, our results demonstrated that BIRC5 might affect immune cell infiltration, making them a predictive biomarker for immunotherapy in LGG patients. Finally, we found that BIRC5 was correlated with sensitivity to diverse drugs in the cancer therapeutic response portal database, with some positive and some negative correlations (Figure 9). For the biological function on the glioma cells, we found that BIRC5 was highly expressed in glioma cell lines, depletion of BIRC5 significantly inhibiting the migration of glioma cells (Figure 10).
In summary, we found that BIRC5 was significantly aberrantly overexpressed in glioma tissues and cells. Through a series of comprehensive approaches, we demonstrated that the upregulated BIRC5 expression is strongly associated with clinicopathologic features, adverse clinical outcomes, and immune cell infiltration in low-grdae glioma. Altogether, our results suggest that BIRC5 might serve as a valuable prognostic factor and a promising novel immunotherapy target for glioma.
This study improves our understanding of the correlation between BIRC5 and low-grade glioma, but some limitations still exist. First, although we explored the correlation between BIRC5 and immune infiltration in LGG patients, there is a lack of experiments to validation the function of BIRC5 in the tumor microenvironment regulation of LGG. Second, we uncover that depletion of BIRC5 was inhibits cell migration of LGG cells. However, the potential molecular mechanisms of BIRC5 in tumor metastasis need to be explored in further studies. Third, we did not conduct the in vivo experiments to validation the function of BIRC5 in the tumor metastasis and tumor microenvironment regulation of LGG. In the future, we will pay more attention to the function of BIRC5 in tumor metastasis and tumor microenvironment regulation of LGG. Furthermore, we will perform more in vivo and vitro experiments to explore the function and the potential molecular mechanisms of BIRC5 in tumor metastasis and tumor microenvironment regulation of LGG.
Overall, our results confirmed that BIRC5 could serve as a potential novel prognostic biomarker for LGG. Moreover, we explored the underlying evidence indicating that BIRC5 regulates immune cell infiltration in the tumor microenvironment in LGG patients. Therefore, these findings are potentially valuable in advancing our current understanding of not only the role of BIRC5 but also its translational use in LGG prognosis and immunotherapy.
Conclusion
Our study found that BIRC5 expression was increased in low-grade glioma, which was also associated with poor prognosis. Furthermore, low-grade glioma might be involved in the initiation and progression of LGG by regulating the function of immune infiltrating cells and cell migration. Herein, we revealed the biological functions of BIRC5 in LGG and offered a potential strategy for the diagnosis and treatment of LGG.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Author Contributions
XJ, YS, and XC designed this work, performed related assay. HX analyzed data. LL and JP supervised and wrote the manuscript. All authors have read and approved the final version of the manuscript.
Funding
This work was supported by National Nature Science Foundation of China (82160512) and Yunnan Applied Basic Research Projects (2017FE467 and 2018FE001).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors would like to thank the support from the department of Neurosurgery, The Second Affiliated Hospital of Kunming Medical University, Kunming 650223, China.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmolb.2022.773662/full#supplementary-material
References
Aldape, K., Brindle, K. M., Chesler, L., Chopra, R., Gajjar, A., Gilbert, M. R., et al. (2019). Challenges to Curing Primary Brain Tumours. Nat. Rev. Clin. Oncol. 16, 509–520. doi:10.1038/s41571-019-0177-5
Basu, A., Bodycombe, N. E., Cheah, J. H., Price, E. V., Liu, K., Schaefer, G. I., et al. (2013). An Interactive Resource to Identify Cancer Genetic and Lineage Dependencies Targeted by Small Molecules. Cell 154, 1151–1161. doi:10.1016/j.cell.2013.08.003
Cao, L., Li, C., Shen, S., Yan, Y., Ji, W., Wang, J., et al. (2013). OCT4 Increases BIRC5 and CCND1 Expression and Promotes Cancer Progression in Hepatocellular Carcinoma. BMC cancer 13, 82. doi:10.1186/1471-2407-13-82
Chai, R.-C., Wu, F., Wang, Q.-X., Zhang, S., Zhang, K.-N., Liu, Y.-Q., et al. (2019). m6A RNA Methylation Regulators Contribute to Malignant Progression and Have Clinical Prognostic Impact in Gliomas. Aging 11, 1204–1225. doi:10.18632/aging.101829
Chen, Y. G., Chen, R., Ahmad, S., Verma, R., Kasturi, S. P., Amaya, L., et al. (2019). N6-Methyladenosine Modification Controls Circular RNA Immunity. Mol. Cel 76, 96–109. doi:10.1016/j.molcel.2019.07.016
Chuwa, A. H., Sone, K., Oda, K., Ikeda, Y., Fukuda, T., Wada-Hiraike, O., et al. (2016). Significance of Survivin as a Prognostic Factor and a Therapeutic Target in Endometrial Cancer. Gynecol. Oncol. 141, 564–569. doi:10.1016/j.ygyno.2016.04.003
Dai, J. B., Zhu, B., Lin, W. J., Gao, H. Y., Dai, H., Zheng, L., et al. (2020). Identification of Prognostic Significance of BIRC5 in Breast Cancer Using Integrative Bioinformatics Analysis. Biosci. Rep. 40. doi:10.1042/BSR20193678
Hou, J., Zhang, H., Liu, J., Zhao, Z., Wang, J., Lu, Z., et al. (2019). YTHDF2 Reduction Fuels Inflammation and Vascular Abnormalization in Hepatocellular Carcinoma. Mol. Cancer 18, 163. doi:10.1186/s12943-019-1082-3
Huang, X., Zhang, F., He, D., Ji, X., Gao, J., Liu, W., et al. (2021). Immune-Related Gene SERPINE1 Is a Novel Biomarker for Diffuse Lower-Grade Gliomas via Large-Scale Analysis. Front. Oncol. 11, 646060. doi:10.3389/fonc.2021.646060
Jaiswal, P. K., Goel, A., and Mittal, R. D. (2015). Survivin: A Molecular Biomarker in Cancer. Indian J. Med. Res. 141, 389–397. doi:10.4103/0971-5916.159250
Jha, K., Shukla, M., and Pandey, M. (2012). Survivin Expression and Targeting in Breast Cancer. Surg. Oncol. 21, 125–131. doi:10.1016/j.suronc.2011.01.001
LaCasse, E. C., Baird, S., Korneluk, R. G., and MacKenzie, A. E. (1998). The Inhibitors of Apoptosis (IAPs) and Their Emerging Role in Cancer. Oncogene 17, 3247–3259. doi:10.1038/sj.onc.1202569
Li, F., Ambrosini, G., Chu, E. Y., Plescia, J., Tognin, S., Marchisio, P. C., et al. (1998). Control of Apoptosis and Mitotic Spindle Checkpoint by Survivin. Nature 396, 580–584. doi:10.1038/25141
Li, T., Fan, J., Wang, B., Traugh, N., Chen, Q., Liu, J. S., et al. (2017). TIMER: A Web Server for Comprehensive Analysis of Tumor-Infiltrating Immune Cells. Cancer Res. 77, e108–e110. doi:10.1158/0008-5472.can-17-0307
Li, Y., Ge, D., and Lu, C. (2019). The SMART App: an Interactive Web Application for Comprehensive DNA Methylation Analysis and Visualization. Epigenetics & Chromatin 12, 71. doi:10.1186/s13072-019-0316-3
Liang, J., Zhao, W., Tong, P., Li, P., Zhao, Y., Li, H., et al. (2020). Comprehensive Molecular Characterization of Inhibitors of Apoptosis Proteins (IAPs) for Therapeutic Targeting in Cancer. BMC Med. Genomics 13, 7. doi:10.1186/s12920-020-0661-x
Lin, T.-Y., Chan, H.-H., Chen, S.-H., Sarvagalla, S., Chen, P.-S., Coumar, M. S., et al. (2020). BIRC5/Survivin Is a Novel ATG12-ATG5 Conjugate Interactor and an Autophagy-Induced DNA Damage Suppressor in Human Cancer and Mouse Embryonic Fibroblast Cells. Autophagy 16, 1296–1313. doi:10.1080/15548627.2019.1671643
Makuch-Kocka, A., Kocki, J., Brzozowska, A., Bogucki, J., Kołodziej, P., Płachno, B. J., et al. (2021). The BIRC Family Genes Expression in Patients with Triple Negative Breast Cancer. Int. J. Mol. Sci. 22. doi:10.3390/ijms22041820
Miroshnikova, Y. A., Mouw, J. K., Barnes, J. M., Pickup, M. W., Lakins, J. N., Kim, Y., et al. (2016). Tissue Mechanics Promote IDH1-dependent HIF1α-Tenascin C Feedback to Regulate Glioblastoma Aggression. Nat. Cel Biol 18, 1336–1345. doi:10.1038/ncb3429
Modhukur, V., Iljasenko, T., Metsalu, T., Lokk, K., Laisk-Podar, T., and Vilo, J. (2018). MethSurv: a Web Tool to Perform Multivariable Survival Analysis Using DNA Methylation Data. Epigenomics 10, 277–288. doi:10.2217/epi-2017-0118
Ni, K., Sun, G. Z., and Sun, G. (2019). The Identification of Key Biomarkers in Patients with Lung Adenocarcinoma Based on Bioinformatics. Math. biosciences Eng. : MBE 16, 7671–7687. doi:10.3934/mbe.2019384
Ostrom, Q. T., Bauchet, L., Davis, F. G., Deltour, I., Fisher, J. L., Langer, C. E., et al. (2014). The Epidemiology of Glioma in Adults: a "state of the Science" Review. Neuro-oncology 16, 896–913. doi:10.1093/neuonc/nou087
Pu, W., Zheng, Y., and Peng, Y. (2020). Prolyl Isomerase Pin1 in Human Cancer: Function, Mechanism, and Significance. Front. Cel Dev. Biol. 8, 168. doi:10.3389/fcell.2020.00168
Ru, B., Wong, C. N., Tong, Y., Zhong, J. Y., Zhong, S. S. W., Wu, W. C., et al. (2019). TISIDB: an Integrated Repository portal for Tumor-Immune System Interactions. Bioinformatics (Oxford, England) 35, 4200–4202. doi:10.1093/bioinformatics/btz210
Wan, B., Liu, B., Yu, G., Huang, Y., and Lv, C. (2019). Differentially Expressed Autophagy-Related Genes Are Potential Prognostic and Diagnostic Biomarkers in clear-cell Renal Cell Carcinoma. Aging 11, 9025–9042. doi:10.18632/aging.102368
Wang, C., Zheng, X., Shen, C., and Shi, Y. (2012). MicroRNA-203 Suppresses Cell Proliferation and Migration by Targeting BIRC5 and LASP1 in Human Triple-Negative Breast Cancer Cells. J. Exp. Clin. Cancer Res. 31, 58. doi:10.1186/1756-9966-31-58
Wang, L., Huang, J., Jiang, M., and Sun, L. (2011). Survivin (BIRC5) Cell Cycle Computational Network in Human No-Tumor Hepatitis/cirrhosis and Hepatocellular Carcinoma Transformation. J. Cel. Biochem. 112, 1286–1294. doi:10.1002/jcb.23030
Woroniecka, K., Chongsathidkiet, P., Rhodin, K., Kemeny, H., Dechant, C., Farber, S. H., et al. (2018). T-cell Exhaustion Signatures Vary with Tumor Type and Are Severe in Glioblastoma. Clin. Cancer Res. 24, 4175–4186. doi:10.1158/1078-0432.ccr-17-1846
Wu, T., and Dai, Y. (2017). Tumor Microenvironment and Therapeutic Response. Cancer Lett. 387, 61–68. doi:10.1016/j.canlet.2016.01.043
Xiong, Q., Jiang, L., Liu, K., Jiang, X., Liu, B., Shi, Y., et al. (2021). miR-133b Targets NCAPH to Promote β-catenin Degradation and Reduce Cancer Stem Cell Maintenance in Non-small Cell Lung Cancer. Sig Transduct Target. Ther. 6, 252. doi:10.1038/s41392-021-00555-x
Xu, P., Jiang, L., Yang, Y., Wu, M., Liu, B., Shi, Y., et al. (2020). PAQR4 Promotes Chemoresistance in Non-small Cell Lung Cancer through Inhibiting Nrf2 Protein Degradation. Theranostics 10, 3767–3778. doi:10.7150/thno.43142
Xue, S., Hu, M., Iyer, V., and Yu, J. (2017). Blocking the PD-1/pd-L1 Pathway in Glioma: a Potential New Treatment Strategy. J. Hematol. Oncol. 10, 81. doi:10.1186/s13045-017-0455-6
Yamamoto, H., Ngan, C. Y., and Monden, M. (2008). Cancer Cells Survive with Survivin. Cancer Sci. 99, 1709–1714. doi:10.1111/j.1349-7006.2008.00870.x
Yamashita, K., Hosoda, K., Nishizawa, N., Katoh, H., and Watanabe, M. (2018). Epigenetic Biomarkers of Promoter DNA Methylation in the new era of Cancer Treatment. Cancer Sci. 109, 3695–3706. doi:10.1111/cas.13812
Yang, W., Soares, J., Greninger, P., Edelman, E. J., Lightfoot, H., Forbes, S., et al. (2013). Genomics of Drug Sensitivity in Cancer (GDSC): a Resource for Therapeutic Biomarker Discovery in Cancer Cells. Nucleic Acids Res. 41, D955–D961. doi:10.1093/nar/gks1111
Yu-Ju Wu, C., Chen, C.-H., Lin, C.-Y., Feng, L.-Y., Lin, Y.-C., Wei, K.-C., et al. (2020). CCL5 of Glioma-Associated Microglia/macrophages Regulates Glioma Migration and Invasion via Calcium-dependent Matrix Metalloproteinase 2. Neuro-oncology 22, 253–266. doi:10.1093/neuonc/noz189
Zhang, C., Huang, S., Zhuang, H., Ruan, S., Zhou, Z., Huang, K., et al. (2020). YTHDF2 Promotes the Liver Cancer Stem Cell Phenotype and Cancer Metastasis by Regulating OCT4 Expression via m6A RNA Methylation. Oncogene 39, 4507–4518. doi:10.1038/s41388-020-1303-7
Keywords: LGG, BIRC5, M6A RNA methylation, cell migration, immune infiltration, drug sensitivity
Citation: Jiang X, Shi Y, Chen X, Xu H, Huang X, Li L and Pu J (2022) The N6-Methylandenosine-Related Gene BIRC5 as a Prognostic Biomarker Correlated With Cell Migration and Immune Cell Infiltrates in Low Grade Glioma. Front. Mol. Biosci. 9:773662. doi: 10.3389/fmolb.2022.773662
Received: 10 September 2021; Accepted: 27 January 2022;
Published: 03 March 2022.
Edited by:
Ismail Hosen, University of Dhaka, BangladeshReviewed by:
Udhaya Kumar S., Vellore Institute of Technology, IndiaShilpita Karmakar, JAX Cancer Center, United States
Copyright © 2022 Jiang, Shi, Chen, Xu, Huang, Li and Pu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lihua Li, lilihua1229@126.com; Jun Pu, pujun303@aliyun.com
†These authors have contributed equally to this work
 Xiulin Jiang
Xiulin Jiang Yulin Shi
Yulin Shi Xi Chen1,4†
Xi Chen1,4†