- 1Institute of Preventive Veterinary Medicine, Sichuan Agricultural University, Chengdu, China
- 2Research Centre of Avian Disease, College of Veterinary Medicine, Sichuan Agricultural University, Chengdu, China
- 3Key Laboratory of Animal Disease and Human Health of Sichuan Province, Chengdu, China
In a previous study, it was shown that Riemerella anatipestifer, a member of Flavobacteriaceae, is naturally competent. However, whether natural competence is universal in Flavobacteriaceae remains unknown. In this study, it was shown for the first time that Riemerella columbina was naturally competent in the laboratory condition; however, Flavobacterium johnsoniae was not naturally competent under the same conditions. The competence of R. columbina was maintained throughout the growth phases, and the transformation frequency was highest during the logarithmic phase. A competition assay revealed that R. columbina preferentially took up its own genomic DNA over heterologous DNA. The natural transformation frequency of R. columbina was significantly increased in GCB medium without peptone or phosphate. Furthermore, natural transformation of R. columbina was inhibited by 0.5 mM EDTA, but could be restored by the addition of CaCl2, MgCl2, ZnCl2, and MnCl2, suggesting that these divalent cations promote the natural transformation of R. columbina. Overall, this study revealed that natural competence is not universal in Flavobacteriaceae members and triggering of competence differs from species to species.
Introduction
Naturally competent bacteria can actively take up naked DNA from their environment and integrate it into the genome, which is called natural transformation (Mell and Redfield, 2014). As one of the three horizontal gene transfer mechanisms, natural transformation facilitates bacterial acquisition of virulence genes and antibiotic-resistant cassettes to help bacteria adapt to the environment (Wiedenbeck and Cohan, 2011; Seitz and Blokesch, 2013a). Natural transformation was first discovered in Streptococcus pneumoniae in 1928 (Griffith, 1928). Currently, at least 83 species have been found to have natural competence (Johnston et al., 2014; Liu et al., 2017).
In Gram-positive and Gram-negative bacteria, there are different mechanism to take up DNA. Naturally competent Gram-negative bacteria, such as Neisseria species and Haemophilus influenzae, use type IV pili (T4P) to take up exogenous double-stranded DNA (dsDNA), in contrast to Helicobacter pylori, which uses a type IV secretion system (T4SS) (Hofreuter et al., 2000), and Campylobacter jejuni, which uses a type II secretion system (T2SS) (Wiesner et al., 2003) to take up exogenous dsDNA. Gram-positive bacteria, such as S. pneumoniae and Bacillus subtilis, use a competence pseudopilus, a structure similar to T4P, to take up dsDNA (Hahn et al., 2005). Once dsDNA is transported across the outer membrane in Gram-negative bacteria or the peptidoglycan layer in Gram-positive bacteria, dsDNA is degraded to single-stranded DNA (ssDNA) and transported through the pore protein ComEC into the cytoplasm (Johnston et al., 2014). Internalized ssDNA is presumably bound by DNA-processing protein A (DprA), which recruits the recombinase RecA to mediate homologous recombination by facilitating strand exchange (Johnston et al., 2014; Huang et al., 2019). At present, the natural transformation of H. influenzae in Pasteurellaceae, Vibrio cholerae in Vibrionaceae, and S. pneumoniae in Streptococcaceae are well studied (Redfield et al., 2006; Blokesch, 2012; Johnston et al., 2014). Among the six genera of Pasteurellaceae, only the genera Actinobacillus and Haemophilus are naturally competent (Redfield et al., 2006). In Streptococcaceae, both Streptococcus and Lactococcus show natural competence (Griffith, 1928; Dalia et al., 2017). Within the genus Haemophilus, when H. influenzae is transferred from rich medium to defined competence medium (M-IV) or the cell culture reaches stationary phase, it becomes naturally competent (Herriott et al., 1970; Redfield, 1991). However, natural transformation of Haemophilus parasuis was readily induced by nutrient-rich medium (Zhang et al., 2015; Li et al., 2016). All information suggests that the occurrence of natural transformation is different among bacteria, even within the same genus.
In a previous study, it was shown that one member of the Flavobacteriaceae family, Riemerella anatipestifer (R. anatipestifer, RA), which causes septicemic diseases in ducks, geese, turkeys, and other birds (Huang et al., 2017), is naturally competent (Liu et al., 2017). However, whether other Flavobacteriaceae species are also naturally competent and under which condition natural transformation is induced remains unknown. Here, Flavobacterium johnsoniae (F. johnsoniae), a common soil and aquatic bacterium (McBride, 2004), and Riemerella columbina (R. columbina), widely distributed species among pigeon populations (Rubbenstroth et al., 2013), were selected as models to explore the occurrence of natural transformation and its influencing factors.
Materials and Methods
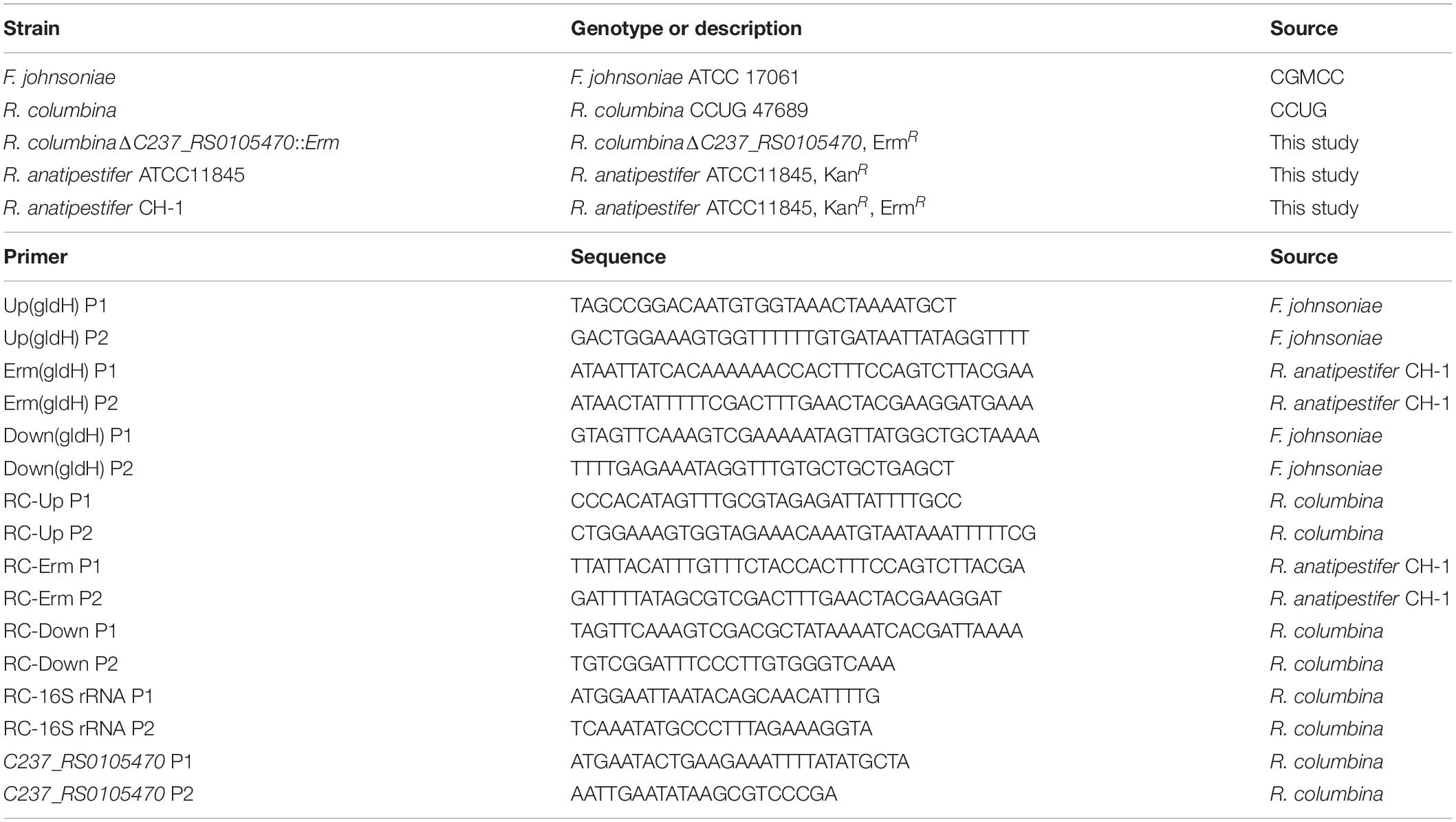
Bacterial Strains, Primers, and Growth Conditions
Riemerella columbina and Flavobacterium johnsoniae were purchased from the Culture Collection of the University of Gothenburg (CCUG) and the China General Microbiological Culture Collection Center (CGMCC), respectively. The bacterial strains and primers used in this study are listed in Table 1. The culture conditions for R. columbina and F. johnsoniae were identical to those used for R. anatipestifer described in a previous study (Huang et al., 2019). Briefly, R. columbina was cultured in GC broth (GCB) medium with shaking or GCB agar plates and LB plates supplemented with 5% sheep blood (blood plates) at 37°C, however, F. johnsoniae was cultured in GCB medium with shaking or GCB plates at 25°C. When required, erythromycin was added into the medium at a final concentration of 1 μg/ml for R. columbina and 50 μg/ml for F. johnsoniae.
Preparation of Donor DNA
The homologous gene of dprA (C237_RS0105470) in R. columbina, which protects ssDNA and loads RecA to facilitate homologous recombination (Mirouze et al., 2013), and the gliding motility gene gldH in F. johnsoniae were selected as targeted deletion gene, since they are not essential for the growth of bacteria and can be deleted (McBride et al., 2003; Hovland et al., 2017; Huang et al., 2019). Donor DNA was composed of upstream of target gene, an antibiotic resistance cassette and downstream of target gene. Briefly, the ∼620 bp upstream sequence and ∼620 bp downstream sequence of C237_RS0105470 were amplified from the genome of R. columbina using the primers RC-Up P1 and RC-Up P2, RC-Down P1 and RC-Down P2, respectively. The ∼620 bp upstream sequence and ∼620 bp downstream sequence of gldH were amplified from the genome of F. johnsoniae using the primers Up(gldH) P1 and Up(gldH) P2, Down(gldH) P1 and Down(gldH) P2, respectively. An erythromycin resistance cassette was amplified from the genome of R. anatipestifer CH-1 using the primers RC-Erm P1 and RC-Erm P2 or Erm(gldH) P1 and Erm(gldH) P2, respectively (Liao et al., 2015; Luo et al., 2015). The three fragments were fused using overlapping PCR (Xiong et al., 2006; Huang et al., 2017, 2019). The fused fragments served as donor DNA for natural transformation.
Natural Transformation Procedure
The procedure of natural transformation was similar to that used for R. anatipestifer described in a previous study (Liu et al., 2017; Huang et al., 2019). Briefly, R. columbina and F. johnsoniae were cultured in GCB liquid with shaking at 37°C for R. columbina and at 25°C for F. johnsoniae. The bacteria were collected during the logarithmic phase (OD600 = 3–4 for R. columbina, OD600 = 1–1.5 for F. johnsoniae) and adjusted to an optical density (OD) of 1. The growth curve of F. johnsoniae in GCB was shown in Supplementary Figure 1. The donor DNA was added to the bacterial cells and incubated for 1 h at 37°C for R. columbina and at 25°C for F. johnsoniae. Then, 100 μl of cells were plated on GCB agar plates supplemented with erythromycin (1 μg/ml for R. columbina; 50 μg/ml for F. johnsoniae) to count transformants. Then, 10 μl of cells were serially diluted with PBS and plated on GCB agar plates to count viable bacteria. The transformation frequency (TF) was calculated as transformants divided by viable bacteria. Then, 100 μl of cells were plated on GCB supplemented with the corresponding concentration of erythromycin to check for spontaneous mutants.
Determination of Growth Curves
The bacteria were streaked on blood plates or GCB agar plates. A single colony was cultured in 5 ml of GCB liquid medium with shaking at 37°C for 14 h. The bacterial cells were transferred into 20 ml of GCB with or without peptone, phosphate or iron at an OD of 0.05 and cultured at 37°C with shaking. The OD600 was determined every 2 h for 14 h, and natural transformation was performed at the corresponding times.
The Effect of Components of GCB on Natural Transformation in R. columbina
Bacterial cells were cultured to the logarithmic phase (OD600 = 3–4) and adjusted to an OD600 of 1. The bacterial cells were collected and resuspended in GCB medium depleted of vitamin B1 (VB1), glucose, L-glutamine, NaCl, peptone, or phosphate. After the bacteria were incubated at 37°C for 30 min, donor DNA was added to the cultured cells, and natural transformation was performed. Iron is essential for the growth of most bacteria (Liao et al., 2016). To investigate whether iron affects the growth and natural transformation of R. columbina, the growth curve of R. columbina in GCB supplemented with different concentrations of iron chelator ethylenediamine-N,N’-bis(2-hydroxyphenylacetic acid) (EDDHA) according to the method mentioned previously (Press et al., 2001; Liu et al., 2016, 2019) and natural transformation were performed after the bacteria were incubated into GCB supplemented with the corresponding concentration of EDDHA at 37°C for 30 min. The viable bacteria and transformants were counted, and the TF was calculated.
EDTA Treatment
Bacterial cells were cultured until the logarithmic phase (OD600 = 3–4) at 37°C with shaking and adjusted to an OD600 of 1. Three hundred microliters of bacteria were collected and resuspended in GCB medium supplemented with 0.5 mM EDTA. Natural transformation was performed after the bacteria were incubated at 37°C for 30 min. The TF was calculated according to the method described previously. To investigate which divalent cation affects the natural transformation of R. columbina, different concentrations of CaCl2, MgCl2, ZnCl2, MnCl2, or CuCl2 were added into the GCB medium supplemented with the corresponding concentration of EDTA. The bacterial cells were first incubated in the above medium at 37°C for 30 min, and natural transformation was then performed. The TF was calculated as described previously.
Statistics
Statistical analysis was performed using GraphPad Prism 8.0 (GraphPad Software Inc., La Jolla, United States). An unpaired two-tailed Student’s t-test was used to compare two groups, and a value of P < 0.05 was considered significant. Data represent the mean and standard deviation (SD) from at least three independent experiments.
Results
R. columbina, but Not F. johnsoniae, Is Naturally Competent Under the Same Conditions
To assay whether other members of Flavobacteriaceae were able to undergo natural transformation, R. columbina and F. johnsoniae were selected. We used the same method as described in a previous study for R. anatipestifer to determine the natural competence of these two species (Liu et al., 2017). After R. columbina incubated with donor DNA which contains the upstream sequence of C237_RS0105470, an erythromycin resistance cassette and the downstream sequence of C237_RS0105470 (Figure 1A), many resistant colonies grew on the plate with erythromycin. However, no resistant colonies appeared in the control group without donor DNA (the spontaneous mutation rate of erythromycin resistance was lower than the detection limitation). Random single colonies were verified using PCR to ensure that the target gene was replaced by the erythromycin resistance cassette through homologous recombination. As shown in Figure 1B, compared to the wild-type strain, the resistant colonies contained an erythromycin resistance gene but not a target gene. It was suggested that the target gene has been replaced by the erythromycin resistance cassette and that the target sequence of the R. columbina strain was lost. It was strongly supported that R. columbina was naturally competent and that natural transformation could be used to efficiently generate targeted gene disruptions in R. columbina, with a TF of 4.14 (±0.5) × 10–6.
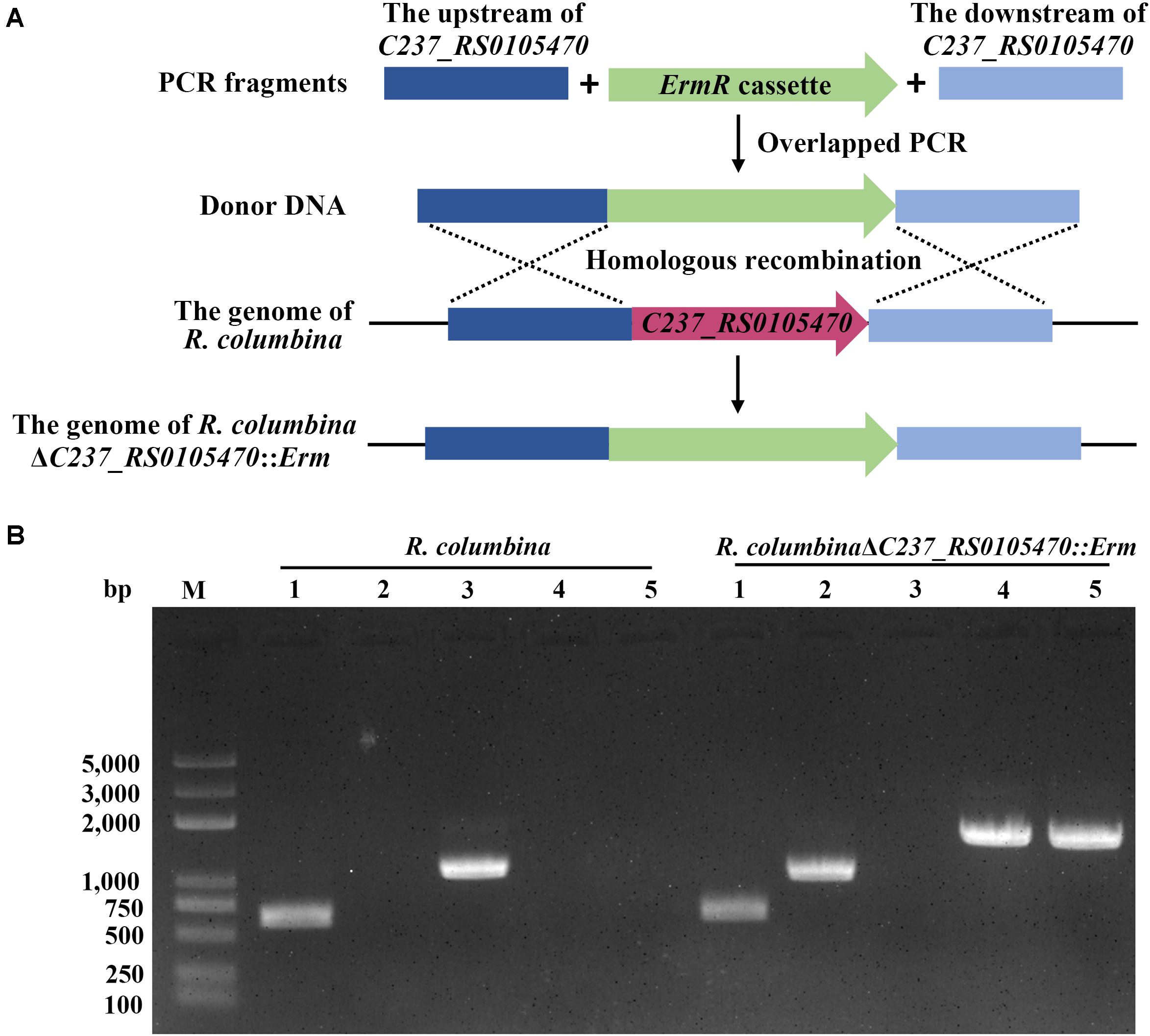
Figure 1. Verification of the mutant strain R. columbinaΔC237_RS0105470::Erm. (A) Schematic diagram of donor DNA preparation and gene deletion based on natural transformation. (B) The identification of the R. columbinaΔC237_RS0105470:Erm mutant strain by PCR. Lane M, MD103 DNA marker (BioMed, Beijing, China). Lane 1, the 16S rRNA was amplified from wild-type R. columbina and the mutant strain R. columbinaΔC237_RS0105470::Erm using the primers RC-16S rRNA P1 and RC-16S rRNA P2. Lane 2, the erythromycin resistance cassette was amplified from wild-type R. columbina and the mutant strain R. columbinaΔC237_RS0105470::Erm using the primers RC-Erm P1 and RC-Erm P2. Lane 3, the target gene C237_RS0105470 was amplified from wild-type R. columbina and the mutant strain R. columbinaΔC237_RS0105470::Erm using the primers C237_RS0105470 P1 and C237_RS0105470 P2. Lane 4, the upstream sequence and erythromycin resistance cassette was amplified from wild-type R. columbina and the mutant strain R. columbinaΔC237_RS0105470::Erm using the primers RC-Up P1 and RC-Erm P2. Lane 5, the erythromycin resistance cassette and downstream sequence was amplified from wild-type R. columbina and the mutant strain R. columbinaΔC237_RS0105470::Erm using the primers RC-Erm P1 and RC-Down P2.
After F. johnsoniae incubated with donor DNA containing the upstream sequence of gldH, an erythromycin resistance cassette and the downstream sequence of gldH, the transformants were selected on GCB plates supplemented with 50 μg/ml erythromycin (the MIC of erythromycin for F. johnsoniae is 16 μg/ml). However, no resistant colony appeared with or without donor DNA. It has been shown that gldH can be deleted in F. johnsoniae through other methods (McBride et al., 2003), suggesting that this gene is not essential for the survival of the bacteria. Overall, it was suggested that F. johnsoniae could not perform natural transformation using the same method as R. columbina under the same conditions.
Searching for the Components of the Natural Transformation Machinery in the Genome of F. johnsoniae
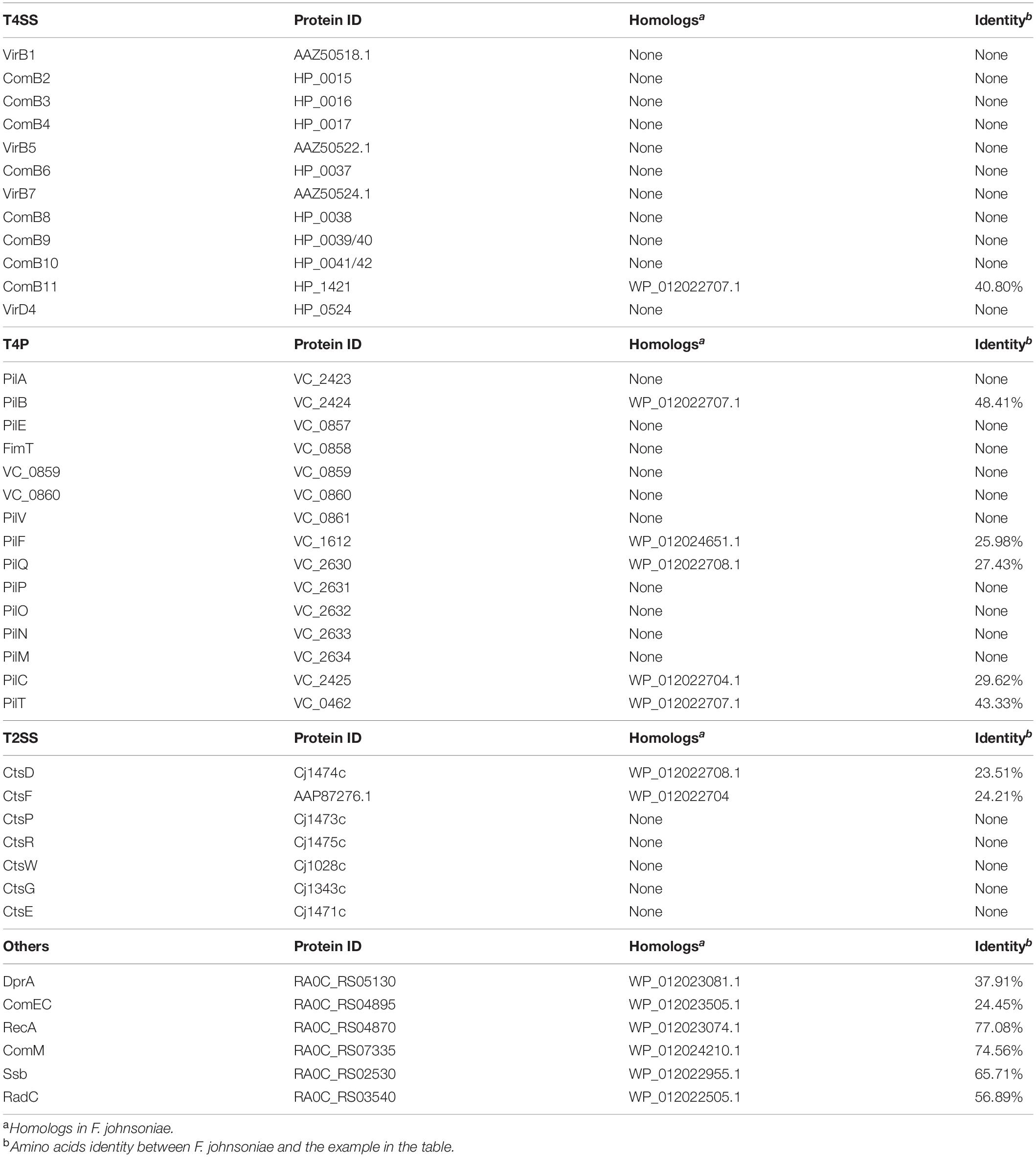
To investigate whether F. johnsoniae contains the homologous proteins that involved in natural transformation. We aligned the amino acids sequences of T4SS from H. pylori and Agrobacterium tumefaciens, T4P from V. cholerae, T2SS from C. jejuni, and other hypothetical competence proteins from R. anatipestifer with genome of F. johnsoniae. As shown in Table 2, only the homolog of ComB11 in T4SS, which is a putative VirB11-homologous ATPase (Karnholz et al., 2006), was found in F. johnsoniae and showed 40.8% identity with the ComB11 of H. pylori. Based on the T4P of V. cholerae (Seitz and Blokesch, 2013b), only the homologs of PilB, PilC, PilF, PilQ and PilT were discovered in F. johnsoniae and shared 48.41, 29.62, 25.98, 27.43, and 43.33% with each relative protein of V. cholerae, respectively. PilB and PilT are polymerization and depolymerization ATPases, respectively (Seitz and Blokesch, 2013b). PilC was an inner membrane platform protein which interacts with PilB and PilT to control both pilus assembly and disassembly (Takhar et al., 2013). PilF is pilolin protein which is essential for pilus biogenesis (Matthey and Blokesch, 2016). PilQ is a secretion pore, which plays a role in translocating pilus on the cell surface (Wolfgang et al., 2000). Furthermore, only the homologs of CtsD and CtsF were found in F. johnsoniae based on the T2SS of C. jejuni (Wiesner et al., 2003). CtsD is an outer membrane protein which has homology to the PilQ protein (Wiesner et al., 2003). CtsF is an inner membrane protein and shares similarity to PilG of N. gonorrhoeae which has homology to the PilC of V. cholerae (Tønjum et al., 1995). Other hypothetical competence protein of R. anatipestifer, like DprA, ComEC, RecA, Ssb, ComM and RadC is also present in F. johnsoniae (Liu et al., 2017). These results indicated that these homologs of F. johnsoniae may be sufficient to encode a T4P-type DNA uptake system in addition to the proteins usually needed for DNA translocation and cytoplasmic processing.
Natural Transformation of R. columbina Increases During the Logarithmic Phase
We were wondering whether natural transformation was able to occur in all growth phases in R. columbina, the TF was assayed. Natural transformation was performed at each time point by adding the same amount of donor DNA. As shown in Figure 2, natural formation of R. columbina occurred in all growth phases, and the TF was the highest during the logarithmic phase [TF = 6.45 (±0.55) × 10–6] and lowest in the lag phase [TF = 6.35 (±0.5) × 10–8]. The number of transformants in different growth phases were included in the Supplementary Data Sheet 2. To investigate the saturated concentration of donor DNA for logarithmic growth period bacteria, R. columbina was cultured to the logarithmic phase and mixed with different amounts of donor DNA (0.1, 1, 10, 100, 200, 500, 1,000, 2,000, or 4,000 ng). As shown in Figure 3, the TF increased with increasing DNA concentration when the amount was lower than 1,000 ng. However, when the DNA amount was higher than 1,000 ng, the TF no longer increased. The number of transformants were included in the Supplementary Data Sheet 2. These results suggested that 1,000 ng of donor DNA was saturating for natural transformation in R. columbina.
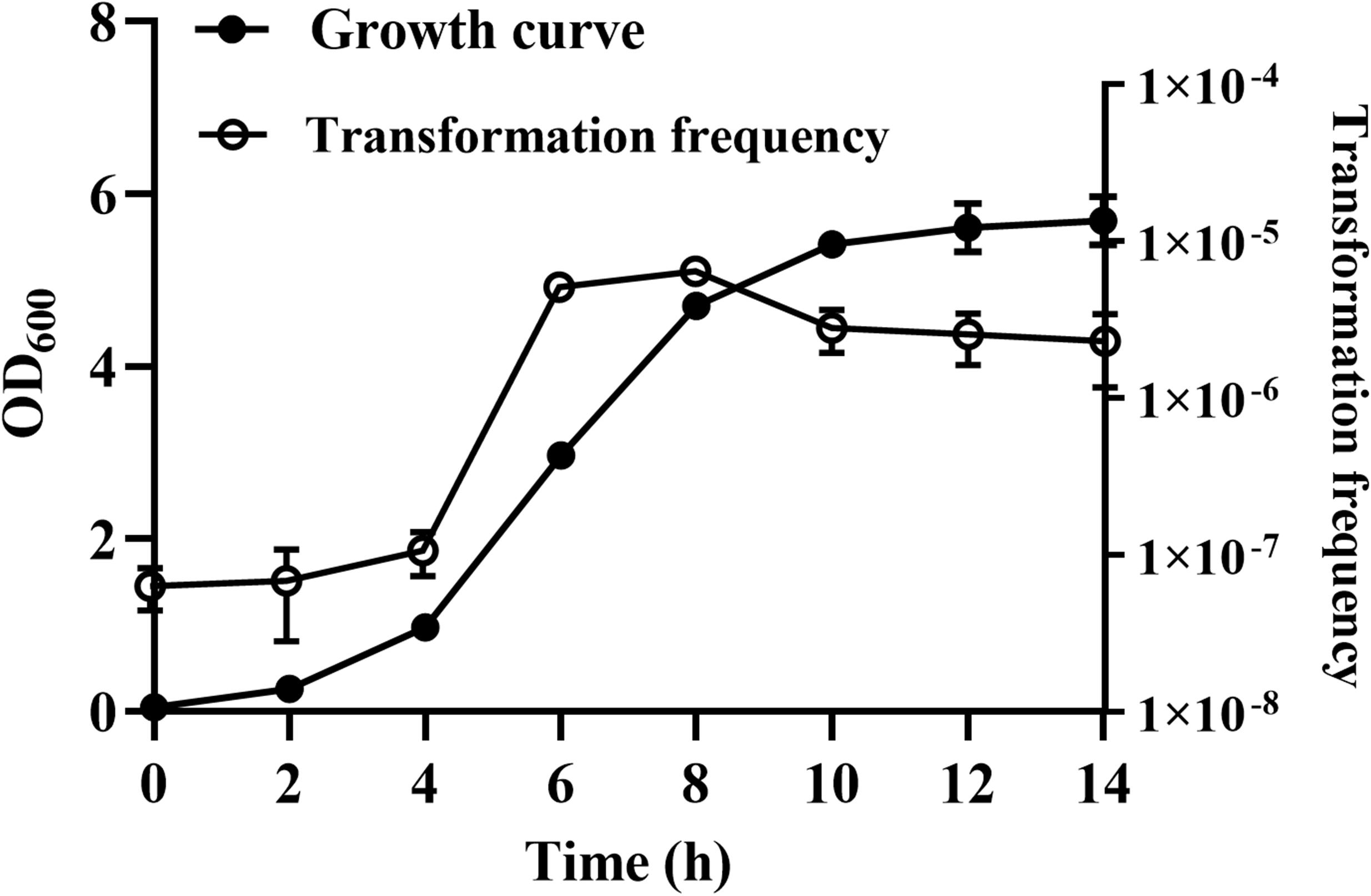
Figure 2. The effect of different growth phases on natural transformation. R. columbina was cultured with shaking at an OD600 of 0.05 for 14 h. The OD600 value was determined every 2 h. Additionally, natural transformation was assessed every 2 h. The results are representative of three independent experiments. Error bars denote standard deviation.
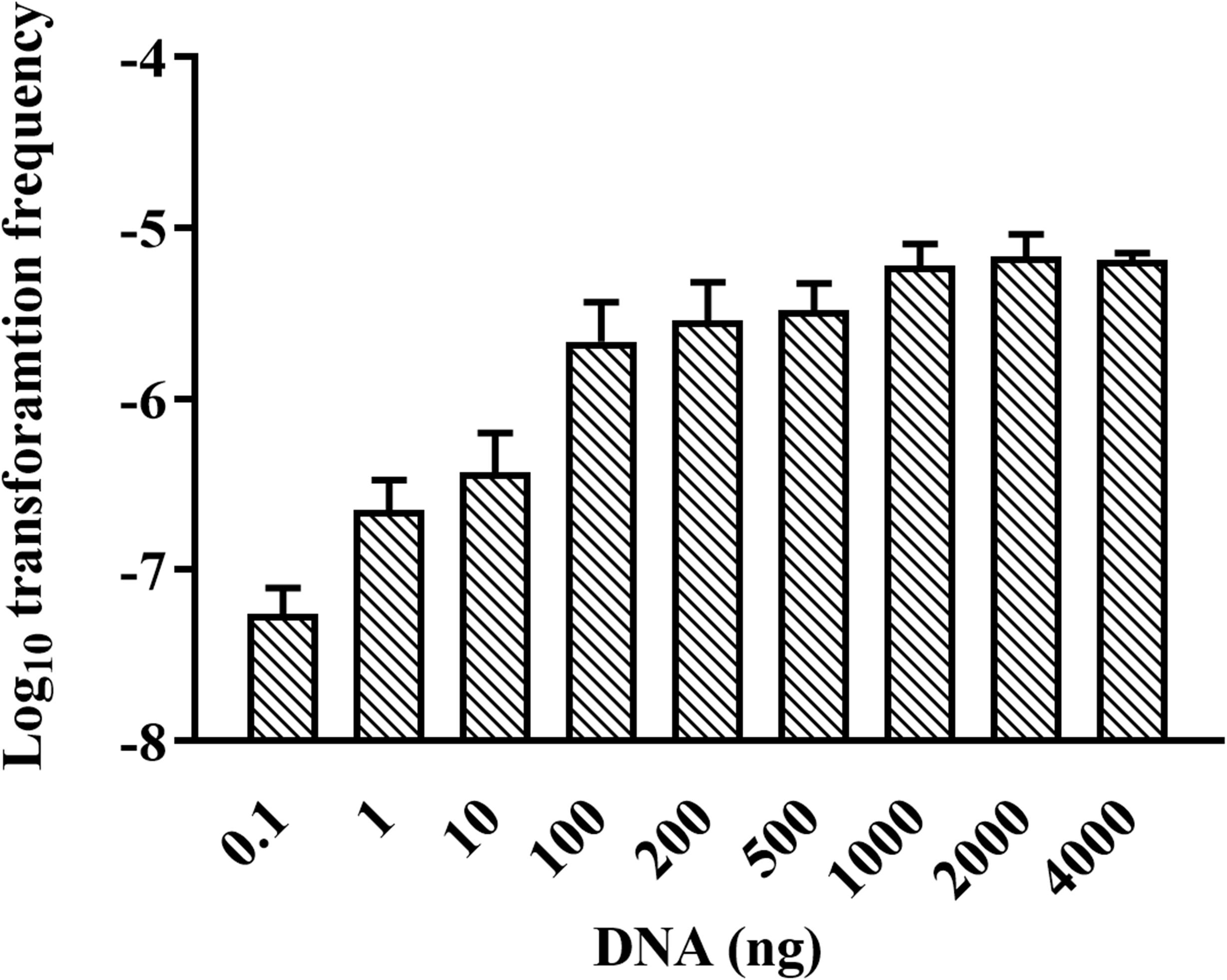
Figure 3. Effect of donor DNA amount on the TF of R. columbina. Donor DNA (0.1 to 4,000 ng) was added to 300 μl of cell cultures at an OD600 of 1 and incubated at 37°C for 1 h. The viable bacteria and erythromycin-resistant transformants were counted. The TF was calculated as transformants divided by the number of viable bacteria. All the results are representative of three independent experiments. Error bars denote standard deviation.
R. columbina Preferentially Takes Up Its Own DNA Over Heterologous DNA
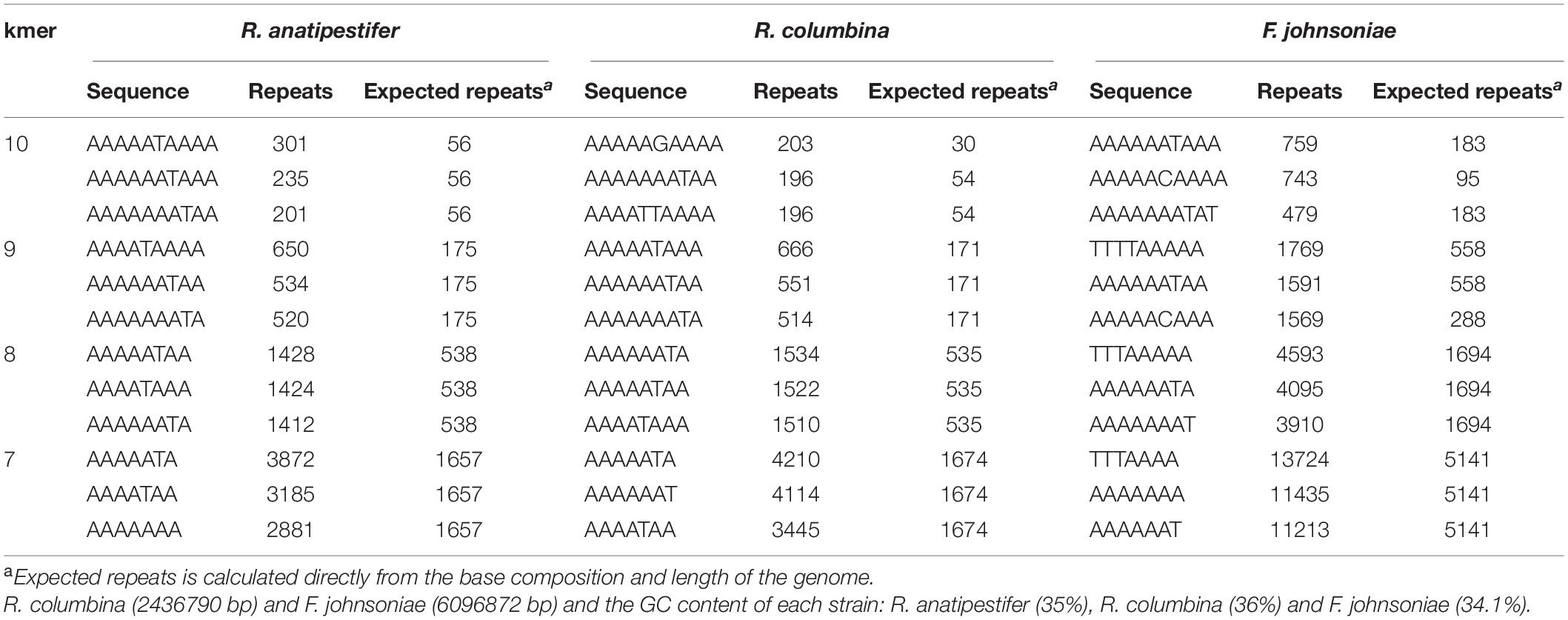
It has been reported that some bacteria, such as H. influenzae and Neisseria, preferentially take up DNA containing short motifs known as uptake signal sequences (USSs) or DNA uptake sequences (DUSs) (Scocca et al., 1974; Sisco and Smith, 1979). These short motifs have accumulated in the genome to high densities over evolutionary time (Mell and Redfield, 2014). To determine whether R. columbina also preferentially takes up its own DNA, a natural transformation competition experiment was performed. In this experiment, the genome of R. columbina ΔC237_RS0105470 was used as the donor DNA. As shown in Figure 4, when 1 μg of donor DNA was mixed with 1 μg of genomic DNA of R. columbina, the TF was decreased two-fold compared to the control in which without competition DNA was added; Moreover, the TF was decreased with the increase of competing DNA. However, the TF showed no significant changes when 1 μg donor of DNA was mixed with 1 μg of R. anatipestifer or E. coli genomic DNA compared to that when only 1 μg of donor DNA was added. The TF was decreased significantly only when the R. anatipestifer or E. coli genomic DNA was increased to 10 μg, which can be considered as unspecific effect. The number of transformants were included in the Supplementary Data Sheet 2. To further investigate whether R. columbina, F. johnsoniae or R. anatipestifer contain putative DUSs or USSs, Jellyfish1 was to be used to count the numbers of occurrences of individual kmers in both strands of their genome, respectively, with a parameter that limited the length of kmers to less than 10 bp. As shown in Table 3, sequences with the top three repeats for 10 bp, 9 bp, 8 bp, and 7 bp were listed, respectively. It was found that hundreds of repeat sequences or its complement were present in their genomes. The frequency of the 9-bp repeat sequence is 0.6/kb for R. anatipestifer, 0.5/kb for R. columbina and 0.5/kb for F. johnsoniae, respectively, which is much higher than the frequency of 0.1/kb expected for a random sequence of this base composition for them. Whether this sequence has the function of DUSs or USSs needs to be further investigated.
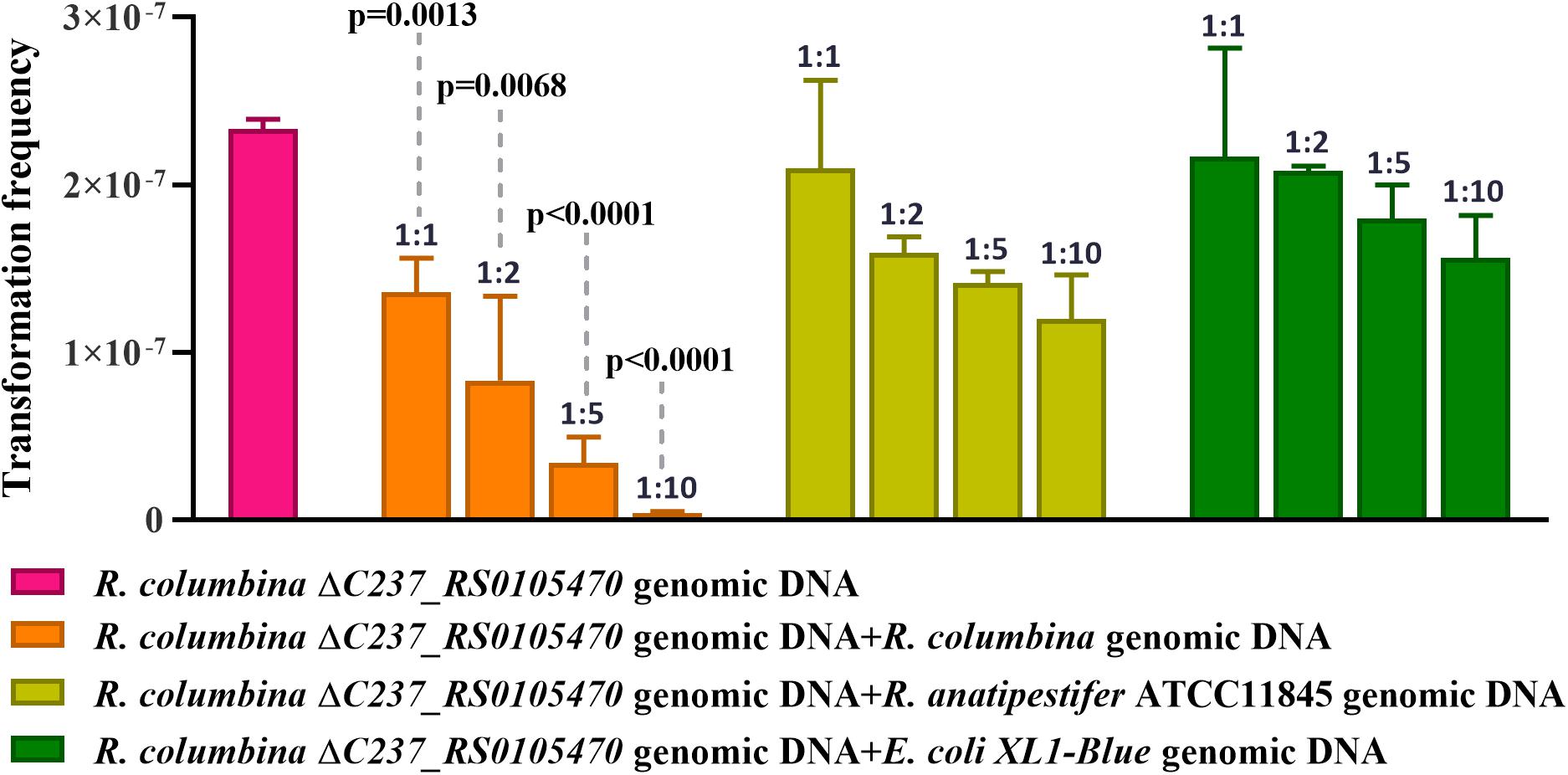
Figure 4. Natural transformation competition experiments. R. columbina was cultured to the logarithmic phase (3–4), and the OD600 value was determined. The cell culture was adjusted to an OD600 of 1. A 300 μl cell culture was transformed with 1 μg the genome of R. columbina ΔC237_RS0105470 (control) or 1 μg of donor DNA mixed with different quantity of competing genomic DNA of R. columbina, R. anatipestifer ATCC11845 or E. coli XL1-Blue and incubated at 37°C for 1 h. The natural TF was calculated as transformants divided by viable bacteria. The P-value refers to the difference between the no-competition and competition measurement, respectively. Insignificance p-value (P > 0.05) was not shown. All the results are representative of three independent experiments. Error bars denote standard deviation.
The TF of R. columbina Is Increased Under Peptone-Restrictive or Phosphate-Restrictive Conditions
To investigate the effect of the nutrients on natural transformation, natural transformation was conducted in GCB depleted for each component, including vitamin B1 (VB1), glucose, L-glutamine, NaCl, peptone and phosphate. As shown in Figure 5A, the TF of R. columbina was 1.9 (±0.1) × 10–5 in GCB depleted of peptone, which increased five-fold compared to that in GCB. The TF of R. columbina was 9.05 (±0.5) × 10–6 in GCB depleted of phosphate, which increased approximately two-fold compared to that in GCB. However, compared to the TF of R. columbina in GCB, there was no significant difference when VB1, glucose, L-glutamine or NaCl was removed from GCB (Figure 5A). The number of transformants were included in the Supplementary Data Sheet 2.
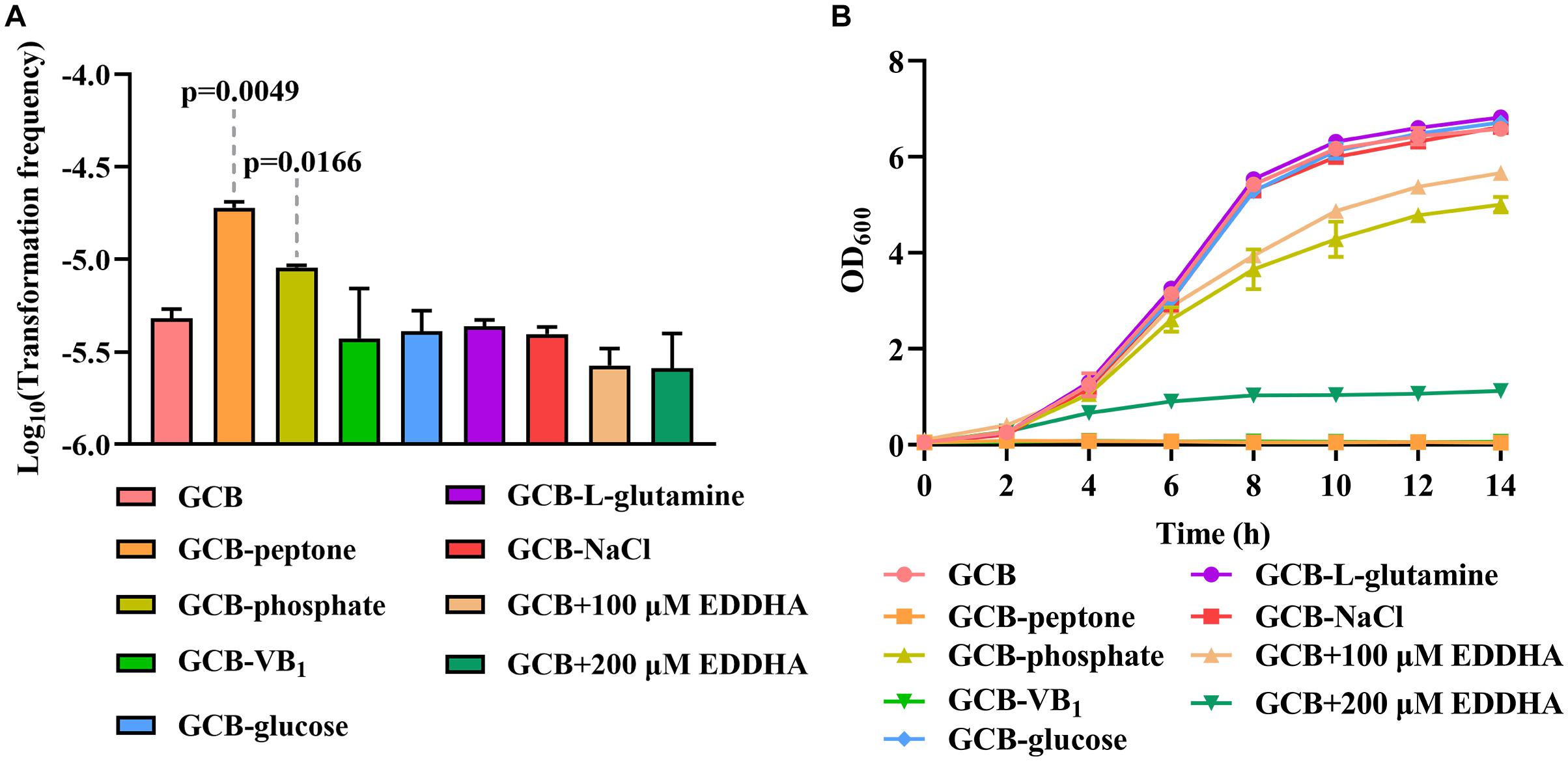
Figure 5. The effect of GCB ingredients on natural transformation and growth in R. columbina. (A) TFs of R. columbina in GCB, GCB depleted of VB1, glucose, L-glutamine, NaCl, peptone or phosphate and GCB supplemented with different concentrations of EDDHA. The P-value refers to the difference between the TF in GCB and in GCB depleted of peptone or phosphate, respectively. Insignificance p-value (P > 0.05) was not shown. (B) Growth curves of R. columbina in GCB, GCB depleted of VB1, glucose, L-glutamine, NaCl, peptone, or phosphate and GCB supplemented with different concentrations of EDDHA. The bacteria were cultured in 20 ml of the above medium inoculated at an OD600 of 0.05, and the OD600 value was determined every 2 h. All the results are representative of three independent experiments. Error bars denote standard deviation.
Next, we investigated whether the change in TF was associated with the growth ability of bacteria. Therefore, the growth curve of bacteria was determined when peptone, phosphate, NaCl, glucose, L-glutamine or VB1 was removed from GCB. The results showed that bacteria did not grow in GCB without peptone or VB1, whereas the growth of bacteria was significantly decreased in GCB without phosphate; however, there were no significant differences when NaCl, glucose, or L-glutamine was removed from GCB (Figure 5B). To investigate whether iron affects the natural transformation of R. columbina, different concentrations of iron chelator EDDHA were supplemented into the GCB medium. As shown in Figure 5A, the TF did not change compared to that of the control (without EDDHA). The number of transformants were included in the Supplementary Data Sheet 2. However, the growth of R. columbina was significantly inhibited in iron-depleted medium (GCB supplemented with 200 μM EDDHA), suggesting that iron is essential for the growth of R. columbina (Figure 5B). Overall, these results suggested that peptone-restrictive or phosphate-restrictive medium had an effect on the natural transformation and this effect is not directly related to the growth ability.
Ca2+, Mg2+, Zn2+, and Mn2+ but Not Cu2+ Promote the Natural Transformation of R. columbina
Iron has no effect on natural transformation, and we wondered if other divalent cations influence the natural transformation of R. columbina. Natural transformation was conducted in GCB medium with 0.5 mM EDTA, which had no effect on the survival of bacteria (Supplementary Figure 2). The results showed that the TF in GCB with 0.5 mM EDTA was 5.75 (±0.75) × 10–7, which was decreased approximately 4-fold compared to that in GCB [TF = 2.5(±0.1) × 10–6], suggesting that 0.5 mM EDTA had a significant inhibitory effect on natural transformation in R. columbina (Figure 6A). To investigate which divalent cation has an effect on natural transformation, different concentrations of CaCl2, MgCl2, ZnCl2, MnCl2, or CuCl2 were supplemented into the cell culture after incubation with EDTA. The addition of 0.5 mM Ca2+ basically restored transformation, and the TF increased as the concentration of Ca2+ increased (Figure 6B). The addition of 0.5 mM Mg2+ completely restored the TF; however, the frequency did not increase as the concentration of Mg2+ increased (Figure 6B), suggesting that 0.5 mM was likely a saturating concentration of Mg2+ for natural transformation in R. columbina. The TF gradually increased as the concentration of Zn2+ increased from 0.1 mM to 0.5 mM. The TF was the highest at 0.5 mM Mn2+(Figure 6B). However, the addition of different concentrations of Cu2+ did not restore the natural transformation but instead inhibited natural transformation (Figure 6B). The number of transformants were included in the Supplementary Data Sheet 2. Therefore, it was shown that Ca2+, Mg2+, Zn2+, and Mn2+ were required for the natural transformation of R. columbina, but Cu2+ was not.
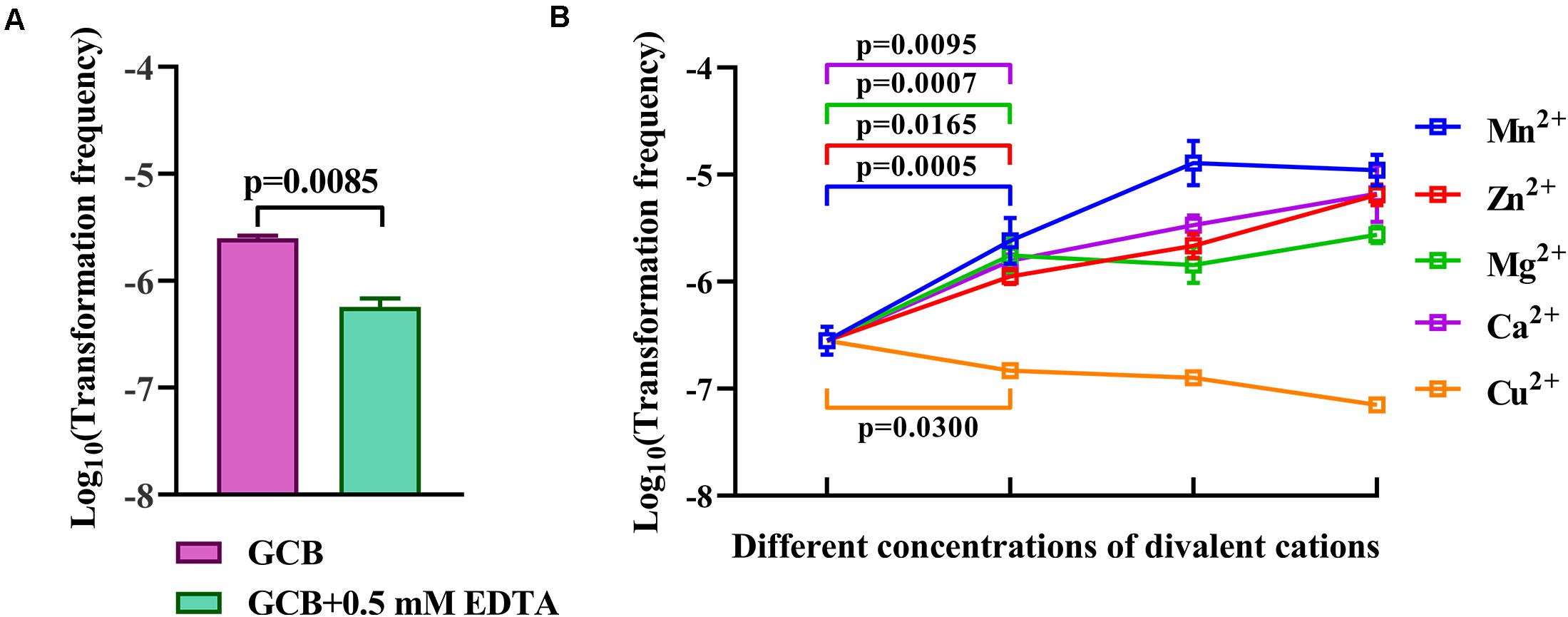
Figure 6. Effect of EDTA and different divalent cations on the natural transformation of R. columbina. (A) Viable bacteria were counted after treatment with or without 0.5 mM EDTA for 1 h, and the effect of 0.5 mM EDTA on the natural transformation of R. columbina was determined. The P-value refers to the difference between the TF in GCB and in GCB containing 0.5 mM EDTA. (B) The natural TF of R. columbina in GCB supplemented with 0.5 mM EDTA and different supplemented concentrations of divalent cation. The concentration of Ca2+ are 0.5, 1, and 5 mM, respectively. The concentration of Mg2+ are 0.5, 1, and 5 mM, respectively. The concentration of Zn2+ are 0.1, 0.2, and 0.5 mM, respectively. The concentration of Mn2+ are 0.2, 0.5, and 1 mM, respectively. The concentration of Cu2+ are 0.1, 0.2, and 0.5 mM, respectively. The P-value refers to the difference between the TF in GCB containing 0.5 mM EDTA and GCB containing 0.5 mM EDTA supplemented by Ca2+, Mg2+, Mg2+, or Cu2+, respectively. All the results are representative of three independent experiments. Error bars denote standard deviation.
Discussion
Riemerella anatipestifer is the first bacterium of Flavobacteriaceae to be reported to have natural competence (Liu et al., 2017). To check whether other bacteria in Flavobacteriaceae are also naturally competent, F. johnsoniae and R. columbina were selected. The results showed that R. columbina was able to undergo natural transformation; however, F. johnsoniae was not competent under the same conditions. One possibility is that the natural transformation of F. johnsoniae does not occur at all growth phases but only at a certain time point, for example natural transformation happens to S. pneumoniae, in which natural transformation is not constitutive, as synthesis and assembly of the uptake apparatus is a transient and regulated process (Mirouze et al., 2013). Another possibility is that the PCR fragments are not suitable substrates, such as occurs with C. jejuni, which takes up only methylated DNA but not PCR fragments (Beauchamp et al., 2017), and H. influenzae and Neisseria gonorrhoeae, which preferentially take up DNA with an USS or DUS over other sources of DNA (Mell et al., 2012; Berry et al., 2013; Frye et al., 2013; Mell and Redfield, 2014). The third possibility is that natural transformation in F. johnsoniae must be induced by special substrates, such as occurs with natural transformation of Vibrio cholerae, which is induced by chitin (Meibom et al., 2005). The fourth possibility is that we did not choose the correct isolates. It has been reported that even for the competent bacteria, some isolates are non-transformable (Evans and Rozen, 2013; Dalia et al., 2015). The last possibility is that F. johnsoniae does not undergo natural transformation because of the lack of some essential genes for natural transformation.
Consistent with the natural transformation of R. anatipestifer (Liu et al., 2017), the natural transformation of R. columbina is also constitutive, although the TF is different at the different growth phases. This phenomenon might occur because the expression of genes involved in natural transformation in R. columbina is different at the different growth phases. Similar to R. anatipestifer, R. columbina preferentially takes up self-sourced genomic DNA, suggesting that each bacterium might use a certain mechanism, such as a restriction modification (R-M) system (Aras et al., 2002; Zhang and Blaser, 2012) or other systems, to prevent the uptake of excessive extracellular DNA that may overload the bacteria, subverting the bacterial genome with extracellular DNA from competing strains.
Originally, the function of natural transformation was hypothesized as “DNA for food” (Redfield, 2001), because the natural competence of H. influenzae and B. subtilis was activated under nutrient-limited condition (Bobb, 1963; Herriott et al., 1970). However, this hypothesis is questionable, since the natural competence of some other bacteria, such as A. baumannii, requires a nutrient-rich condition (Traglia et al., 2016). In the case of R. columbina, we showed that the TF of R. columbina was significantly increased under peptone-restrictive or phosphate-restrictive conditions, suggesting that the uptake of DNA may be “food” for R. columbina to supplement the nitrogen and phosphorus.
A more plausible hypothesis for the function of natural transformation is “DNA for repair”(Michod et al., 2008; Engelmoer et al., 2013), since the natural transformation of some bacteria, such as H. pylori (Dorer et al., 2010), S. pneumoniae (Prudhomme et al., 2006) and B. subtilis (Zhang et al., 2018), was activated by antibiotics or DNA damage reagent. Here, we also investigated the effects of antibiotics such as ampicillin (inhibitor of cell wall biosynthesis), kanamycin (inhibitor of protein biosynthesis), nalidixic acid (inhibitor of DNA replication) and mitomycin C (intercalation with DNA) during the natural transformation of R. columbina. We showed that none of the antibiotics affected natural TF of R. columbina did not change after treatment with antibiotics (Supplementary Figure 3), suggesting that the antibiotics used here cannot trigger natural transformation of R. columbina.
Our systematic investigation of natural transformation in the Flavobacteriaceae family shows that it is widely distributed. However, the environmental conditions that trigger natural transformation vary from species to species. In this family, natural transformation appears to play a major role in HGT. The discovery of natural transformation in R. columbina represents the basis for the establishment of gene editing and cloning system in this bacterium.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Author Contributions
ML, DZ, and AC conceived and designed the experiments. LH, LX, MH, CX, SZ, QG, DS, and BT performed the experiments. MW, RJ, SC, XZ, QY, and YW analyzed the data. JH, XO, and SM contributed to reagents, materials, and analysis tools. ML, FB, and AC wrote the manuscript. All authors have reviewed the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (32072825), China Agricultural Research System (CARS-42-17), Sichuan Veterinary Medicine and Drug Innovation Group of China Agricultural Research System (SCCXTD-2020-18), and Integration and Demonstration of Key Technologies for Goose Industrial Chain in Sichuan Province (2018NZ0005).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.634895/full#supplementary-material
Footnotes
References
Aras, R. A., Small, A. J., Ando, T., and Blaser, M. J. (2002). Helicobacter pylori interstrain restriction-modification diversity prevents genome subversion by chromosomal DNA from competing strains. Nucleic Acids Res. 30, 5391–5397. doi: 10.1093/nar/gkf686
Beauchamp, J. M., Leveque, R. M., Dawid, S., and DiRita, V. J. (2017). Methylation-dependent DNA discrimination in natural transformation of Campylobacter jejuni. Proc. Natl. Acad. Sci. U.S.A. 114, E8053–E8061. doi: 10.1073/pnas.1703331114
Berry, J. L., Cehovin, A., McDowell, M. A., Lea, S. M., and Pelicic, V. (2013). Functional analysis of the interdependence between DNA uptake sequence and its cognate ComP receptor during natural transformation in Neisseria species. PLoS Genet. 9:e1004014. doi: 10.1371/journal.pgen.1004014
Blokesch, M. (2012). Chitin colonization, chitin degradation and chitin-induced natural competence of Vibrio cholerae are subject to catabolite repression. Environ. Microbiol. 14, 1898–1912. doi: 10.1111/j.1462-2920.2011.02689.x
Bobb, D. (1963). Overnight incubation technique for obtaining transformable Bacillus subtilus cells of reproducible competency. Nature 199, 828–829. doi: 10.1038/199828a0
Dalia, A. B., Seed, K. D., Calderwood, S. B., and Camilli, A. (2015). A globally distributed mobile genetic element inhibits natural transformation of Vibrio cholerae. Proc. Natl. Acad. Sci. U.S.A. 112, 10485–10490. doi: 10.1073/pnas.1509097112
Dalia, T. N., Yoon, S. H., Galli, E., Barre, F. X., Waters, C. M., and Dalia, A. B. (2017). Enhancing multiplex genome editing by natural transformation (MuGENT) via inactivation of ssDNA exonucleases. Nucleic Acids Res. 45, 7527–7537. doi: 10.1093/nar/gkx496
Dorer, M. S., Fero, J., and Salama, N. R. (2010). DNA damage triggers genetic exchange in Helicobacter pylori. PLoS Pathog. 6:e1001026. doi: 10.1371/journal.ppat.1001026
Engelmoer, D. J., Donaldson, I., and Rozen, D. E. (2013). Conservative sex and the benefits of transformation in Streptococcus pneumoniae. PLoS Pathog. 9:e1003758. doi: 10.1371/journal.ppat.1003758
Evans, B. A., and Rozen, D. E. (2013). Significant variation in transformation frequency in Streptococcus pneumoniae. ISME J. 7, 791–799. doi: 10.1038/ismej.2012.170
Frye, S. A., Nilsen, M., Tonjum, T., and Ambur, O. H. (2013). Dialects of the DNA uptake sequence in Neisseriaceae. PLoS Genet. 9:e1003458. doi: 10.1371/journal.pgen.1003458
Griffith, F. (1928). The significance of pneumococcal types. J. Hyg. 27, 113–159. doi: 10.1017/s0022172400031879
Hahn, J., Maier, B., Haijema, B. J., Sheetz, M., and Dubnau, D. (2005). Transformation proteins and DNA uptake localize to the cell poles in Bacillus subtilis. Cell 122, 59–71. doi: 10.1016/j.cell.2005.04.035
Herriott, R. M., Meyer, E. M., and Vogt, M. (1970). Defined nongrowth media for stage II development of competence in Haemophilus influenzae. J. Bacteriol. 101, 517–524. doi: 10.1128/jb.101.2.517-524.1970
Hofreuter, D., Odenbreit, S., Puls, J., Schwan, D., and Haas, R. (2000). Genetic competence in Helicobacter pylori: mechanisms and biological implications. Res. Microbiol. 151, 487–491. doi: 10.1016/s0923-2508(00)00164-9
Hovland, E., Beyene, G. T., Frye, S. A., Homberset, H., Balasingham, S. V., Gomez-Munoz, M., et al. (2017). DprA from Neisseria meningitidis: properties and role in natural competence for transformation. Microbiology 163, 1016–1029. doi: 10.1099/mic.0.000489
Huang, L., Tian, X., Liu, M., Wang, M., Biville, F., Cheng, A., et al. (2019). DprA is essential for natural competence in riemerella anatipestifer and has a conserved evolutionary mechanism. Front. Genet. 10:429. doi: 10.3389/fgene.2019.00429
Huang, L., Yuan, H., Liu, M. F., Zhao, X. X., Wang, M. S., Jia, R. Y., et al. (2017). Type B chloramphenicol acetyltransferases are responsible for chloramphenicol resistance in Riemerella anatipestifer, China. Front. Microbiol. 8:297. doi: 10.3389/fmicb.2017.00297
Johnston, C., Martin, B., Fichant, G., Polard, P., and Claverys, J. P. (2014). Bacterial transformation: distribution, shared mechanisms and divergent control. Nat. Rev. Microbiol. 12, 181–196. doi: 10.1038/nrmicro3199
Karnholz, A., Hoefler, C., Odenbreit, S., Fischer, W., Hofreuter, D., and Haas, R. (2006). Functional and topological characterization of novel components of the comB DNA transformation competence system in Helicobacter pylori. J. Bacteriol. 188, 882–893. doi: 10.1128/jb.188.3.882-893.2006
Li, J., Yuan, X., Xu, L., Kang, L., Jiang, J., and Wang, Y. (2016). Efficient construction of Haemophilus parasuis mutants based on natural transformation. Can. J. Vet. Res. 80, 281–286.
Liao, H., Cheng, X., Zhu, D., Wang, M., Jia, R., Chen, S., et al. (2015). TonB energy transduction systems of Riemerella anatipestifer are required for iron and hemin utilization. PLoS One 10:e0127506. doi: 10.1371/journal.pone.0127506
Liao, H., Liu, M., Cheng, X., Zhu, D., Wang, M., Jia, R., et al. (2016). The detection of hemin-binding proteins in Riemerella anatipestifer CH-1. Curr. Microbiol. 72, 152–158. doi: 10.1007/s00284-015-0932-5
Liu, M., Huang, M., Huang, L., Biville, F., Zhu, D., Wang, M., et al. (2019). New perspectives on Galleria mellonella larvae as a host model using Riemerella anatipestifer as a proof of concept. Infect. Immun. 87:e00072-19. doi: 10.1128/IAI.00072-19
Liu, M., Wang, M., Zhu, D., Wang, M., Jia, R., Chen, S., et al. (2016). Investigation of TbfA in Riemerella anatipestifer using plasmid-based methods for gene over-expression and knockdown. Sci. Rep. 6:37159. doi: 10.1038/srep37159
Liu, M., Zhang, L., Huang, L., Biville, F., Zhu, D., Wang, M., et al. (2017). Use of natural transformation to establish an easy knockout method in Riemerella anatipestifer. Appl. Environ. Microbiol. 83:e000127-17. doi: 10.1128/AEM.00127-17
Luo, H., Liu, M., Wang, L., Zhou, W., Wang, M., Cheng, A., et al. (2015). Identification of ribosomal RNA methyltransferase gene ermF in Riemerella anatipestifer. Avian Pathol. 44, 162–168. doi: 10.1080/03079457.2015.1019828
Matthey, N., and Blokesch, M. (2016). The DNA-Uptake process of naturally competent Vibrio cholerae. Trends Microbiol. 24, 98–110. doi: 10.1016/j.tim.2015.10.008
McBride, M. J. (2004). Cytophaga-flavobacterium gliding motility. J. Mol. Microbiol. Biotechnol. 7, 63–71. doi: 10.1159/000077870
McBride, M. J., Braun, T. F., and Brust, J. L. (2003). Flavobacterium johnsoniae GldH is a lipoprotein that is required for gliding motility and chitin utilization. J. Bacteriol. 185, 6648–6657. doi: 10.1128/jb.185.22.6648-6657.2003
Meibom, K. L., Blokesch, M., Dolganov, N. A., Wu, C. Y., and Schoolnik, G. K. (2005). Chitin induces natural competence in Vibrio cholerae. Science 310, 1824–1827. doi: 10.1126/science.1120096
Mell, J. C., Hall, I. M., and Redfield, R. J. (2012). Defining the DNA uptake specificity of naturally competent Haemophilus influenzae cells. Nucleic Acids Res. 40, 8536–8549. doi: 10.1093/nar/gks640
Mell, J. C., and Redfield, R. J. (2014). Natural competence and the evolution of DNA uptake specificity. J. Bacteriol. 196, 1471–1483. doi: 10.1128/JB.01293-13
Michod, R. E., Bernstein, H., and Nedelcu, A. M. (2008). Adaptive value of sex in microbial pathogens. Infect. Genet. Evol. 8, 267–285. doi: 10.1016/j.meegid.2008.01.002
Mirouze, N., Berge, M. A., Soulet, A. L., Mortier-Barriere, I., Quentin, Y., Fichant, G., et al. (2013). Direct involvement of DprA, the transformation-dedicated RecA loader, in the shut-off of pneumococcal competence. Proc. Natl. Acad. Sci. U.S.A. 110, E1035–E1044. doi: 10.1073/pnas.1219868110
Press, C. M., Loper, J. E., and Kloepper, J. W. (2001). Role of iron in rhizobacteria-mediated induced systemic resistance of cucumber. Phytopathology 91, 593–598. doi: 10.1094/PHYTO.2001.91.6.593
Prudhomme, M., Attaiech, L., Sanchez, G., Martin, B., and Claverys, J. P. (2006). Antibiotic stress induces genetic transformability in the human pathogen Streptococcus pneumoniae. Science 313, 89–92. doi: 10.1126/science.1127912
Redfield, R. J. (1991). sxy-1, a Haemophilus influenzae mutation causing greatly enhanced spontaneous competence. J. Bacteriol. 173, 5612–5618. doi: 10.1128/jb.173.18.5612-5618.1991
Redfield, R. J., Findlay, W. A., Bosse, J., Kroll, J. S., Cameron, A. D., and Nash, J. H. (2006). Evolution of competence and DNA uptake specificity in the Pasteurellaceae. BMC Evol. Biol. 6:82. doi: 10.1186/1471-2148-6-82
Rubbenstroth, D., Ryll, M., Hotzel, H., Christensen, H., Knobloch, J. K., Rautenschlein, S., et al. (2013). Description of Riemerella columbipharyngis sp. nov., isolated from the pharynx of healthy domestic pigeons (Columba livia f. domestica), and emended descriptions of the genus Riemerella, Riemerella anatipestifer and Riemerella columbina. Int. J. Syst. Evol. Microbiol. 63(Pt 1), 280–287. doi: 10.1099/ijs.0.036798-0
Scocca, J. J., Poland, R. L., and Zoon, K. C. (1974). Specificity in deoxyribonucleic acid uptake by transformable Haemophilus influenzae. J. Bacteriol. 118, 369–373. doi: 10.1128/jb.118.2.369-373.1974
Seitz, P., and Blokesch, M. (2013a). Cues and regulatory pathways involved in natural competence and transformation in pathogenic and environmental Gram-negative bacteria. FEMS Microbiol. Rev. 37, 336–363. doi: 10.1111/j.1574-6976.2012.00353.x
Seitz, P., and Blokesch, M. (2013b). DNA-uptake machinery of naturally competent Vibrio cholerae. Proc. Natl. Acad. Sci. U.S.A. 110, 17987–17992. doi: 10.1073/pnas.1315647110
Sisco, K. L., and Smith, H. O. (1979). Sequence-specific DNA uptake in Haemophilus transformation. Proc. Natl. Acad. Sci. U.S.A. 76, 972–976. doi: 10.1073/pnas.76.2.972
Takhar, H. K., Kemp, K., Kim, M., Howell, P. L., and Burrows, L. L. (2013). The platform protein is essential for type IV pilus biogenesis. J. Biol. Chem. 288, 9721–9728. doi: 10.1074/jbc.m113.453506
Tønjum, T., Freitag, N. E., Namork, E., and Koomey, M. (1995). Identification and characterization of pilG, a highly conserved pilus−assembly gene in pathogenic Neisseria. Mol. Microbiol. 16, 451–464. doi: 10.1111/j.1365-2958.1995.tb02410.x
Traglia, G. M., Quinn, B., Schramm, S. T., Soler-Bistue, A., and Ramirez, M. S. (2016). Serum Albumin and Ca2+ are natural competence inducers in the human pathogen Acinetobacter baumannii. Antimicrob. Agents Chemother. 60, 4920–4929. doi: 10.1128/AAC.00529-16
Wiedenbeck, J., and Cohan, F. M. (2011). Origins of bacterial diversity through horizontal genetic transfer and adaptation to new ecological niches. FEMS Microbiol. Rev. 35, 957–976. doi: 10.1111/j.1574-6976.2011.00292.x
Wiesner, R. S., Hendrixson, D. R., and DiRita, V. J. (2003). Natural transformation of Campylobacter jejuni requires components of a type II secretion system. J. Bacteriol. 185, 5408–5418. doi: 10.1128/jb.185.18.5408-5418.2003
Wolfgang, M., van Putten, J. P., Hayes, S. F., Dorward, D., and Koomey, M. (2000). Components and dynamics of fiber formation define a ubiquitous biogenesis pathway for bacterial pili. Embo J. 19, 6408–6418. doi: 10.1093/emboj/19.23.6408
Xiong, A. S., Yao, Q. H., Peng, R. H., Duan, H., Li, X., Fan, H. Q., et al. (2006). PCR-based accurate synthesis of long DNA sequences. Nat. Protoc. 1, 791–797. doi: 10.1038/nprot.2006.103
Zhang, L., Li, Y., Dai, K., Wen, X., Wu, R., Huang, X., et al. (2015). Establishment of a successive markerless mutation system in Haemophilus parasuis through natural transformation. PLoS One 10:e0127393. doi: 10.1371/journal.pone.0127393
Zhang, X., Jin, T., Deng, L., Wang, C., Zhang, Y., and Chen, X. (2018). Stress-induced, highly efficient, donor cell-dependent cell-to-cell natural transformation in Bacillus subtilis. J. Bacteriol. 200:e00267-18. doi: 10.1128/JB.00267-18
Keywords: Flavobacteriaceae, R. columbina, Flavobacterium johnsoniae, natural competence, horizontal gene transfer
Citation: Huang L, Liu M, Zhu D, Xie L, Huang M, Xiang C, Biville F, Jia R, Chen S, Zhao X, Yang Q, Wu Y, Zhang S, Huang J, Ou X, Mao S, Gao Q, Sun D, Tian B, Wang M and Cheng A (2021) Natural Transformation of Riemerella columbina and Its Determinants. Front. Microbiol. 12:634895. doi: 10.3389/fmicb.2021.634895
Received: 29 November 2020; Accepted: 12 February 2021;
Published: 03 March 2021.
Edited by:
Friedrich Götz, University of Tübingen, GermanyReviewed by:
Rosemary Redfield, University of British Columbia, CanadaYuichi Koga, Osaka University, Japan
Copyright © 2021 Huang, Liu, Zhu, Xie, Huang, Xiang, Biville, Jia, Chen, Zhao, Yang, Wu, Zhang, Huang, Ou, Mao, Gao, Sun, Tian, Wang and Cheng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mingshu Wang, mshwang@163.com; Anchun Cheng, chenganchun@vip.163.com
†These authors have contributed equally to this work
 Li Huang
Li Huang Mafeng Liu
Mafeng Liu Dekang Zhu
Dekang Zhu Li Xie1,2,3
Li Xie1,2,3 Shun Chen
Shun Chen Shaqiu Zhang
Shaqiu Zhang Xumin Ou
Xumin Ou Sai Mao
Sai Mao Bin Tian
Bin Tian Mingshu Wang
Mingshu Wang Anchun Cheng
Anchun Cheng