Abstract
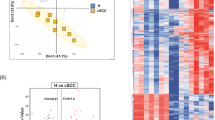
Long noncoding RNAs (lncRNAs) are fundamental regulators of pre- and post-transcriptional gene regulation. Over 35,000 different lncRNAs have been described with some of them being involved in cancer formation. The present study was initiated to describe differentially expressed lncRNAs in basal cell carcinoma (BCC). Patients with BCC (n = 6) were included in this study. Punch biopsies were harvested from the tumor center and nonlesional epidermal skin (NLES, control, n = 6). Microarray-based lncRNA and mRNA expression profiles were identified through screening for 30,586 lncRNAs and 26,109 protein-coding transcripts (mRNAs). The microarray data were validated by RT-PCR in a second set of BCC versus control samples. Gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses of mRNAs were performed to assess biologically relevant pathways. A total of 1851 lncRNAs were identified as being significantly up-regulated, whereas 2165 lncRNAs were identified as being significantly down-regulated compared to nonlesional skin (p < 0.05). Oncogenic and/or epidermis-specific lncRNAs, such as CASC15 or ANRIL, were among the differentially expressed sequences. GO analysis showed that the highest enriched GO targeted by up-regulated transcripts was “extracellular matrix.” KEGG pathway analysis showed the highest enrichment scores in “Focal adhesion.” BCC showed a significantly altered lncRNA and mRNA expression profile. Dysregulation of previously described lncRNAs may play a role in the molecular pathogenesis of BCC and should be subject of further analysis.





Similar content being viewed by others
References
Mattick JS. Non-coding RNAs: the architects of eukaryotic complexity. EMBO Rep. 2001;2:986–91.
Sand M. The pathway of miRNA maturation. Methods Mol Biol. 2014;1095:3–10.
Sand M, Gambichler T, Sand D, Skrygan M, Altmeyer P, Bechara FG. MicroRNAs and the skin: tiny players in the body’s largest organ. J Dermatol Sci. 2009;53:169–75.
Sand M, Skrygan M, Sand D, Georgas D, Hahn SA, Gambichler T, et al. Expression of microRNAs in basal cell carcinoma. Br J Dermatol. 2012;167:847–55.
Sand M, Gambichler T, Skrygan M, Sand D, Scola N, Altmeyer P, et al. Expression levels of the microRNA processing enzymes Drosha and dicer in epithelial skin cancer. Cancer Investig. 2010;28:649–53.
Sand M, Skrygan M, Georgas D, Arenz C, Gambichler T, Sand D, et al. Expression levels of the microRNA maturing microprocessor complex component DGCR8 and the RNA-induced silencing complex (RISC) components argonaute-1, argonaute-2, PACT, TARBP1, and TARBP2 in epithelial skin cancer. Mol Carcinog. 2012;51:916–22.
Sand M, Sand D, Altmeyer P, Bechara FG. MicroRNA in non-melanoma skin cancer. Cancer Biomarkers : Sect Dis Markers. 2012;11:253–7.
Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775–89.
Guo X, Gao L, Wang Y, Chiu DK, Wang T, Deng Y et al. Advances in long noncoding rnas: Identification, structure prediction and function annotation. Brief Funct Genomics 2015, in press
Hombach S, Kretz M. The non-coding skin: exploring the roles of long non-coding RNAs in epidermal homeostasis and disease. Bioessays. 2013;35:1093–100.
Ezkurdia I, Juan D, Rodriguez JM, Frankish A, Diekhans M, Harrow J, et al. Multiple evidence strands suggest that there may be as few as 19 000 human protein-coding genes. Hum Mol Genet. 2014;23:5866–78.
Li X, Wu Z, Fu X, Han W. LncRNAs: insights into their function and mechanics in underlying disorders. Mutat Res Rev Mutat Res. 2014;762:1–21.
Isin M, Dalay N. LncRNAs and neoplasia. Clin Chim Acta. 2015;444:280–8.
Kretz M, Webster DE, Flockhart RJ, Lee CS, Zehnder A, Lopez-Pajares V, et al. Suppression of progenitor differentiation requires the long noncoding RNA ANCR. Genes Dev. 2012;26:338–43.
Lopez-Pajares V, Qu K, Zhang J, Webster DE, Barajas BC, Siprashvili Z, et al. A LncRNA-MAF:MAFB transcription factor network regulates epidermal differentiation. Dev Cell. 2015;32:693–706.
Orom UA, Derrien T, Beringer M, Gumireddy K, Gardini A, Bussotti G, et al. Long noncoding RNAs with enhancer-like function in human cells. Cell. 2010;143:46–58.
Mazar J, Sinha S, Dinger ME, Mattick JS, Perera RJ. Protein-coding and non-coding gene expression analysis in differentiating human keratinocytes using a three-dimensional epidermal equivalent. Mol Genet Genomics : MGG. 2010;284:1–9.
Yarmishyn AA, Kurochkin IV. Long noncoding RNAs: a potential novel class of cancer biomarkers. Front Genet. 2015;6:145.
Hajjari M, Khoshnevisan A, Shin YK. Molecular function and regulation of long non-coding RNAs: paradigms with potential roles in cancer. Tumour Biol. 2014;35:10645–63.
Hsu F, Kent WJ, Clawson H, Kuhn RM, Diekhans M, Haussler D. The UCSC known genes. Bioinformatics. 2006;22:1036–46.
Harrow J, Denoeud F, Frankish A, Reymond A, Chen CK, Chrast J, et al. GENCODE: producing a reference annotation for encode. Genome Biol. 2006;7(1):1–9. S4.
Pang KC, Stephen S, Dinger ME, Engstrom PG, Lenhard B, Mattick JS. RNAdb 2.0-—an expanded database of mammalian non-coding RNAs. Nucleic Acids Res. 2007;35:D178–82.
Dinger ME, Pang KC, Mercer TR, Crowe ML, Grimmond SM, Mattick JS. NRED: a database of long noncoding RNA expression. Nucleic Acids Res. 2009;37:D122–6.
Amaral PP, Clark MB, Gascoigne DK, Dinger ME, Mattick JS. LncRNAdb: A reference database for long noncoding RNAs. Nucleic Acids Res. 2011;39:D146–51.
Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci U S A. 2009;106:11667–72.
Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, et al. Integrative annotation of human large intergenic noncodingRNA reveals global properties and specific subclasses. Genes Dev. 2011;25:1915–27.
Bejerano G, Pheasant M, Makunin I, Stephen S, Kent WJ, Mattick JS, et al. Ultraconserved elements in the human genome. Science. 2004;304:1321–5.
Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–23.
Luo X, Shi Q, Gu Y, Pan J, Hua M, Liu M, et al. LncRNA pathway involved in premature preterm rupture of membrane (PPROM): an epigenomic approach to study the pathogenesis of reproductive disorders. PLoS One. 2013;8, e79897.
Gene Ontology C. Gene ontology consortium: going forward. Nucleic Acids Res. 2015;43:D1049–56.
Consortium TGO. Gene ontology consortium: going forward. Nucleic Acids Res. 2015;43:D1049–56.
Benjamini Y, Drai D, Elmer G, Kafkafi N, Golani I. Controlling the false discovery rate in behavior genetics research. Behav Brain Res. 2001;125:279–84.
Quackenbush J. Computational analysis of microarray data. Nat Rev Genet. 2001;2:418–27.
Sokal RR, Michener CD. A statistical method for evaluating systematic relationships. Univ Kansas Sci Bull. 1958;28:1409–38.
Gene Ontology C. The gene ontology project in 2008. Nucleic Acids Res. 2008;36:D440–4.
Gene Ontology C. The gene ontology in 2010: extensions and refinements. Nucleic Acids Res. 2010;38:D331–5.
Xing Z, Lin A, Li C, Liang K, Wang S, Liu Y, et al. LncRNA directs cooperative epigenetic regulation downstream of chemokine signals. Cell. 2014;159:1110–25.
Miller SJ. Biology of basal cell carcinoma (part I). J Am Acad Dermatol. 1991;24:1–13.
Miller SJ. Biology of basal cell carcinoma (part II). J Am Acad Dermatol. 1991;24:161–75.
Wang Y, Chen W, Chen J, Pan Q, Pan J. LncRNA expression profiles of EGFR exon 19 deletions in lung adenocarcinoma ascertained by using microarray analysis. Med Oncol. 2014;31:137.
Chen X. Predicting lncRNA-disease associations and constructing lncRNA functional similarity network based on the information of miRNA. Sci Rep. 2015;5:13186.
Zhuang M, Gao W, Xu J, Wang P, Shu Y. The long non-coding RNA h19-derived miR-675 modulates human gastric cancer cell proliferation by targeting tumor suppressor RUNX1. Biochem Biophys Res Commun. 2014;448:315–22.
Li H, Yu B, Li J, Su L, Yan M, Zhu Z, et al. Overexpression of lncRNA H19 enhances carcinogenesis and metastasis of gastric cancer. Oncotarget. 2014;5:2318–29.
Wang X, El Naqa IM. Prediction of both conserved and nonconserved microRNA targets in animals. Bioinformatics. 2008;24:325–32.
Ivanovska I, Ball AS, Diaz RL, Magnus JF, Kibukawa M, Schelter JM, et al. MicroRNAs in the miR-106b family regulate p21/CDKN1a and promote cell cycle progression. Mol Cell Biol. 2008;28:2167–74.
Russell MR, Penikis A, Oldridge DA, Alvarez-Dominguez JR, McDaniel L, Diamond M, et al. CASC15-s is a tumor suppressor lncRNA at the 6p22 neuroblastoma susceptibility locus. Cancer Res. 2015;75:3155–66.
Lessard L, Liu M, Marzese DM, Wang H, Chong K, Kawas N et al. The casc15 long intergenic noncoding rna locus is involved in melanoma progression and phenotype switching. J Invest Dermatol 2015;135(10):2464–74.
Rabinowitz YS, Dong L, Wistow G. Gene expression profile studies of human keratoconus cornea for NEIBank: a novel cornea-expressed gene and the absence of transcripts for aquaporin 5. Invest Ophthalmol Vis Sci. 2005;46:1239–46.
Naemura M, Murasaki C, Inoue Y, Okamoto H, Kotake Y. Long noncoding RNA ANRIL regulates proliferation of non-small cell lung cancer and cervical cancer cells. Anticancer Res. 2015;35:5377–82.
Iranpour M, Soudyab M, Geranpayeh L, Mirfakhraie R, Azargashb E, Movafagh A et al. Expression analysis of four long noncoding rnas in breast cancer. Tumour Biol 2015, in press
Kretz M, Siprashvili Z, Chu C, Webster DE, Zehnder A, Qu K, et al. Control of somatic tissue differentiation by the long non-coding RNA TINCR. Nature. 2013;493:231–5.
Wan DC, Wang KC. Long noncoding RNA: significance and potential in skin biology. Cold Spring Harb Perspect Med. 2014;4.
Kretz M. TINCR, staufen1, and cellular differentiation. RNA Biol. 2013;10:1597–601.
Szegedi K, Sonkoly E, Nagy N, Nemeth IB, Bata-Csorgo Z, Kemeny L, et al. The anti-apoptotic protein G1P3 is overexpressed in psoriasis and regulated by the non-coding RNA, PRINS. Exp Dermatol. 2010;19:269–78.
Wang KC, Yang YW, Liu B, Sanyal A, Corces-Zimmerman R, Chen Y, et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011;472:120–4.
Jiang YJ, Bikle DD. LncRNA profiling reveals new mechanism for VDR protection against skin cancer formation. J Steroid Biochem Mol Biol. 2014;144(Pt A):87–90.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflicts of interest
None
Financial disclosure
All authors hereby disclose any commercial associations that may pose or create a conflict of interest with the information presented in this manuscript. The authors report no conflicts of interest. The authors alone are responsible for the content and writing of this paper. Daniel Sand was supported by the Heed Ophthalmic Foundation.
Ethics
This study conformed to local requirements following ethical and investigational committee review, informed consent, and other statutes or regulations regarding the protection of the rights and welfare of human subjects participating in medical research (Ethical Review Board of Ruhr-University Bochum, Germany).
Rights and permissions
About this article
Cite this article
Sand, M., Bechara, F.G., Sand, D. et al. Long-noncoding RNAs in basal cell carcinoma. Tumor Biol. 37, 10595–10608 (2016). https://doi.org/10.1007/s13277-016-4927-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13277-016-4927-z