Abstract
Purpose
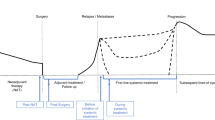
Circulating tumor DNA in plasma may present a minimally invasive opportunity to identify tumor-derived mutations to inform selection of targeted therapies for individual patients, particularly in cases of oligometastatic disease where biopsy of multiple tumors is impractical. To assess the utility of plasma DNA as a “liquid biopsy” for precision oncology, we tested whether sequencing of plasma DNA is a reliable surrogate for sequencing of tumor DNA to identify targetable genetic alterations.
Methods
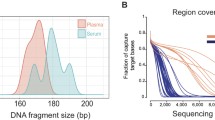
Blood and biopsies of 1–3 tumors were obtained from 4 evaluable patients with advanced breast cancer. One patient provided samples from an additional 7 tumors post-mortem. DNA extracted from plasma, tumor tissues, and buffy coat of blood were used for probe-directed capture of all exons in 149 cancer-related genes and massively parallel sequencing. Somatic mutations in DNA from plasma and tumors were identified by comparison to buffy coat DNA.
Results
Sequencing of plasma DNA identified 27.94 ± 11.81% (mean ± SD) of mutations detected in a tumor(s) from the same patient; such mutations tended to be present at high allelic frequency. The majority of mutations found in plasma DNA were not found in tumor samples. Mutations were also found in plasma that matched clinically undetectable tumors found post-mortem.
Conclusions
The incomplete overlap of genetic alteration profiles of plasma and tumors warrants caution in the sole reliance of plasma DNA to identify therapeutically targetable alterations in patients and indicates that analysis of plasma DNA complements, but does not replace, tumor DNA profiling.
Trial Registration: Subjects were prospectively enrolled in trial NCT01836640 (registered April 22, 2013).





Similar content being viewed by others
Abbreviations
- AF:
-
Allelic frequency
- ctDNA:
-
Cell-free circulating tumor DNA
- FFPE:
-
Formalin-fixed and paraffin-embedded
- GATK:
-
Genome Analysis Toolkit
- [18F]FDG:
-
Fluorodeoxyglucose
- PET:
-
Positron emission tomography
References
Engelman JA, Janne PA, Mermel C, Pearlberg J, Mukohara T, Fleet C, Cichowski K, Johnson BE, Cantley LC (2005) ErbB-3 mediates phosphoinositide 3-kinase activity in gefitinib-sensitive non-small cell lung cancer cell lines. Proc Natl Acad Sci USA 102(10):3788–3793. https://doi.org/10.1073/pnas.0409773102
Martin LA, Pancholi S, Farmer I, Guest S, Ribas R, Weigel MT, Thornhill AM, Ghazoui Z, A'Hern R, Evans DB, Lane HA, Johnston SR, Dowsett M (2012) Effectiveness and molecular interactions of the clinically active mTORC1 inhibitor everolimus in combination with tamoxifen or letrozole in vitro and in vivo. Breast Cancer Res: BCR 14(5):R132. https://doi.org/10.1186/bcr3330
Juric D, Castel P, Griffith M, Griffith OL, Won HH, Ellis H, Ebbesen SH, Ainscough BJ, Ramu A, Iyer G, Shah RH, Huynh T, Mino-Kenudson M, Sgroi D, Isakoff S, Thabet A, Elamine L, Solit DB, Lowe SW, Quadt C, Peters M, Derti A, Schegel R, Huang A, Mardis ER, Berger MF, Baselga J, Scaltriti M (2015) Convergent loss of PTEN leads to clinical resistance to a PI(3)Kalpha inhibitor. Nature 518(7538):240–244. https://doi.org/10.1038/nature13948
Hu ZY, Xie N, Tian C, Yang X, Liu L, Li J, Xiao H, Wu H, Lu J, Gao J, Hu X, Cao M, Shui Z, Xiao M, Tang Y, He Q, Chang L, Xia X, Yi X, Liao Q, Ouyang Q (2018) Identifying circulating tumor DNA mutation profiles in metastatic breast cancer patients with multiline resistance. EBioMedicine 32:111–118. https://doi.org/10.1016/j.ebiom.2018.05.015
Weinberg AL, Carter D, Ahonen M, Alarid ET, Murdoch FE, Fritsch MK (2007) The DNA binding domain of estrogen receptor alpha is required for high-affinity nuclear interaction induced by estradiol. Biochemistry-Us 46(31):8933–8942. https://doi.org/10.1021/bi700018w
Dunn CA, Clark W, Black EJ, Gillespie DAF (2003) Estrogen receptor activation function 2 (AF-2) is essential for hormone-dependent transactivation and cell transformation induced by a v-Jun DNA binding domain-estrogen receptor chimera. Bba-Gene Struct Expr 1628(3):147–155. https://doi.org/10.1016/S0167-4781(03)00136-2
von Lindern M, Boer L, Wessely O, Parker M, Beug H (1998) The transactivation domain AF-2 but not the DNA-binding domain of the estrogen receptor is required to inhibit differentiation of avian erythroid progenitors. Mol Endocrinol 12(2):263–277. https://doi.org/10.1210/Me.12.2.263
Klinge CM, Brolly CL, Bambara RA, Hilf R (1997) hsp70 is not required for high affinity binding of purified calf uterine estrogen receptor to estrogen response element DNA in vitro. J Steroid Biochem 63(4–6):283–301. https://doi.org/10.1016/S0960-0760(97)00091-5
McBride DJ, Orpana AK, Sotiriou C, Joensuu H, Stephens PJ, Mudie LJ, Hamalainen E, Stebbings LA, Andersson LC, Flanagan AM, Durbecq V, Ignatiadis M, Kallioniemi O, Heckman CA, Alitalo K, Edgren H, Futreal PA, Stratton MR, Campbell PJ (2010) Use of cancer-specific genomic rearrangements to quantify disease burden in plasma from patients with solid tumors. Genes Chromosomes Cancer 49(11):1062–1069. https://doi.org/10.1002/gcc.20815
Leary RJ, Kinde I, Diehl F, Schmidt K, Clouser C, Duncan C, Antipova A, Lee C, McKernan K, De La Vega FM, Kinzler KW, Vogelstein B, Diaz LA Jr, Velculescu VE (2010) Development of personalized tumor biomarkers using massively parallel sequencing. Sci Transl Med 2(20):20ra14. https://doi.org/10.1126/scitranslmed.3000702
Sorenson GD, Pribish DM, Valone FH, Memoli VA, Bzik DJ, Yao SL (1994) Soluble normal and mutated DNA sequences from single-copy genes in human blood. Cancer Epidemiol Biomark Prev 3(1):67–71
Garcia-Murillas I, Schiavon G, Weigelt B, Ng C, Hrebien S, Cutts RJ, Cheang M, Osin P, Nerurkar A, Kozarewa I, Garrido JA, Dowsett M, Reis-Filho JS, Smith IE, Turner NC (2015) Mutation tracking in circulating tumor DNA predicts relapse in early breast cancer. Sci Transl Med 7(302):302ra133. https://doi.org/10.1126/scitranslmed.aab0021
Ding L, Ellis MJ, Li S, Larson DE, Chen K, Wallis JW, Harris CC, McLellan MD, Fulton RS, Fulton LL, Abbott RM, Hoog J, Dooling DJ, Koboldt DC, Schmidt H, Kalicki J, Zhang Q, Chen L, Lin L, Wendl MC, McMichael JF, Magrini VJ, Cook L, McGrath SD, Vickery TL, Appelbaum E, Deschryver K, Davies S, Guintoli T, Lin L, Crowder R, Tao Y, Snider JE, Smith SM, Dukes AF, Sanderson GE, Pohl CS, Delehaunty KD, Fronick CC, Pape KA, Reed JS, Robinson JS, Hodges JS, Schierding W, Dees ND, Shen D, Locke DP, Wiechert ME, Eldred JM, Peck JB, Oberkfell BJ, Lolofie JT, Du F, Hawkins AE, O'Laughlin MD, Bernard KE, Cunningham M, Elliott G, Mason MD, Thompson DM Jr, Ivanovich JL, Goodfellow PJ, Perou CM, Weinstock GM, Aft R, Watson M, Ley TJ, Wilson RK, Mardis ER (2010) Genome remodelling in a basal-like breast cancer metastasis and xenograft. Nature 464(7291):999–1005. https://doi.org/10.1038/nature08989
Taniguchi K, Uchida J, Nishino K, Kumagai T, Okuyama T, Okami J, Higashiyama M, Kodama K, Imamura F, Kato K (2011) Quantitative detection of EGFR mutations in circulating tumor DNA derived from lung adenocarcinomas. Clin Cancer Res 17(24):7808–7815. https://doi.org/10.1158/1078-0432.CCR-11-1712
Baselga J, Im S-A, Iwata H, Clemons M, Ito Y, Awada A, Chia S, Jagiello-Gruszfeld A, Pistilli B, Tseng L-M, Hurvitz S, Masuda N, Cortés J, De Laurentiis M, Arteaga CL, Jiang Z, Jonat W, Hachemi S, Le Mouhaër S, Di Tomaso E, Urban P, Massacesi C, Campone M (2015) PIK3CA status in circulating tumor DNA (ctDNA) predicts efficacy of buparlisib (BUP) plus fulvestrant (FULV) in postmenopausal women with endocrine-resistant HR+/HER2– advanced breast cancer (BC): first results from the randomized, phase III BELLE-2 trial. Presented at the San Antonio Breast Cancer Symposium (Dec 11, 2015) Abstract S6-01
Dawson SJ, Tsui DW, Murtaza M, Biggs H, Rueda OM, Chin SF, Dunning MJ, Gale D, Forshew T, Mahler-Araujo B, Rajan S, Humphray S, Becq J, Halsall D, Wallis M, Bentley D, Caldas C, Rosenfeld N (2013) Analysis of circulating tumor DNA to monitor metastatic breast cancer. N Engl J Med 368(13):1199–1209. https://doi.org/10.1056/NEJMoa1213261
Haber DA, Velculescu VE (2014) Blood-based analyses of cancer: circulating tumor cells and circulating tumor DNA. Cancer Discov 4(6):650–661. https://doi.org/10.1158/2159-8290.CD-13-1014
Ignatiadis M, Dawson SJ (2014) Circulating tumor cells and circulating tumor DNA for precision medicine: dream or reality? Ann Oncol 25(12):2304–2313. https://doi.org/10.1093/annonc/mdu480
Smith NG, Gyanchandani R, Shah OS, Gurda GT, Lucas PC, Hartmaier RJ, Brufsky AM, Puhalla S, Bahreini A, Kota K, Wald AI, Nikiforov YE, Nikiforova MN, Oesterreich S, Lee AV (2019) Targeted mutation detection in breast cancer using MammaSeq. Breast Cancer Res 21(1):22. https://doi.org/10.1186/s13058-019-1102-7
Lo YM, Chan KC, Sun H, Chen EZ, Jiang P, Lun FM, Zheng YW, Leung TY, Lau TK, Cantor CR, Chiu RW (2010) Maternal plasma DNA sequencing reveals the genome-wide genetic and mutational profile of the fetus. Sci Transl Med 2(61):61ra91. https://doi.org/10.1126/scitranslmed.3001720
Bussani R, De-Giorgio F, Abbate A, Silvestri F (2007) Cardiac metastases. J Clin Pathol 60(1):27–34. https://doi.org/10.1136/jcp.2005.035105
Bertozzi S, Londero AP, Cedolini C, Uzzau A, Seriau L, Bernardi S, Bacchetti S, Pasqual EM, Risaliti A (2015) Prevalence, risk factors, and prognosis of peritoneal metastasis from breast cancer. SpringerPlus 4:688. https://doi.org/10.1186/s40064-015-1449-x
Maurer AH, Burshteyn M, Adler LP, Steiner RM (2011) How to differentiate benign versus malignant cardiac and paracardiac F-18 FDG uptake at oncologic PET/CT. Radiographics 31(5):1287–1305. https://doi.org/10.1148/rg.315115003
Raptopoulos V, Gourtsoyiannis N (2001) Peritoneal carcinomatosis. Eur Radiol 11(11):2195–2206. https://doi.org/10.1007/s003300100998
Laghi A, Bellini D, Rengo M, Accarpio F, Caruso D, Biacchi D, Di Giorgio A, Sammartino P (2017) Diagnostic performance of computed tomography and magnetic resonance imaging for detecting peritoneal metastases: systematic review and meta-analysis. Radiol Med (Torino) 122(1):1–15. https://doi.org/10.1007/s11547-016-0682-x
Edginton AN, Ritter L (2009) Predicting plasma concentrations of bisphenol A in children younger than 2 years of age after typical feeding schedules, using a physiologically based toxicokinetic model. Environ Health Perspect 117(4):645–652. https://doi.org/10.1289/ehp.0800073
Hay JB, Hobbs BB (1977) The flow of blood to lymph nodes and its relation to lymphocyte traffic and the immune response. J Exp Med 145(1):31–44
Virtanen KA, Lonnroth P, Parkkola R, Peltoniemi P, Asola M, Viljanen T, Tolvanen T, Knuuti J, Ronnemaa T, Huupponen R, Nuutila P (2002) Glucose uptake and perfusion in subcutaneous and visceral adipose tissue during insulin stimulation in nonobese and obese humans. J Clin Endocrinol Metab 87(8):3902–3910. https://doi.org/10.1210/jcem.87.8.8761
Bange J, Prechtl D, Cheburkin Y, Specht K, Harbeck N, Schmitt M, Knyazeva T, Muller S, Gartner S, Sures I, Wang H, Imyanitov E, Haring HU, Knayzev P, Iacobelli S, Hofler H, Ullrich A (2002) Cancer progression and tumor cell motility are associated with the FGFR4 Arg(388) allele. Cancer Res 62(3):840–847
Thussbas C, Nahrig J, Streit S, Bange J, Kriner M, Kates R, Ulm K, Kiechle M, Hoefler H, Ullrich A, Harbeck N (2006) FGFR4 Arg388 allele is associated with resistance to adjuvant therapy in primary breast cancer. J Clin Oncol 24(23):3747–3755. https://doi.org/10.1200/JCO.2005.04.8587
Xu B, Tong N, Chen SQ, Hua LX, Wang ZJ, Zhang ZD, Chen M (2011) FGFR4 Gly388Arg polymorphism contributes to prostate cancer development and progression: a meta-analysis of 2618 cases and 2305 controls. BMC Cancer 11:84. https://doi.org/10.1186/1471-2407-11-84
Hojbjerg JA, Madsen AT, Schmidt HH, Sorensen SF, Stougaard M, Meldgaard P, Sorensen BS (2019) Intra-individual variation of circulating tumour DNA in lung cancer patients. Mol Oncol 13(10):2098–2106. https://doi.org/10.1002/1878-0261.12546
Chae YK, Davis AA, Jain S, Santa-Maria C, Flaum L, Beaubier N, Platanias LC, Gradishar W, Giles FJ, Cristofanilli M (2017) Concordance of genomic alterations by next-generation sequencing in tumor tissue versus circulating tumor DNA in breast cancer. Mol Cancer Ther 16(7):1412–1420. https://doi.org/10.1158/1535-7163.MCT-17-0061
Rothe F, Laes JF, Lambrechts D, Smeets D, Vincent D, Maetens M, Fumagalli D, Michiels S, Drisis S, Moerman C, Detiffe JP, Larsimont D, Awada A, Piccart M, Sotiriou C, Ignatiadis M (2014) Plasma circulating tumor DNA as an alternative to metastatic biopsies for mutational analysis in breast cancer. Ann Oncol 25(10):1959–1965. https://doi.org/10.1093/annonc/mdu288
Frenel JS, Carreira S, Goodall J, Roda D, Perez-Lopez R, Tunariu N, Riisnaes R, Miranda S, Figueiredo I, Nava-Rodrigues D, Smith A, Leux C, Garcia-Murillas I, Ferraldeschi R, Lorente D, Mateo J, Ong M, Yap TA, Banerji U, Tandefelt DG, Turner N, Attard G, de Bono JS (2015) Serial next-generation sequencing of circulating cell-free DNA evaluating tumor clone response to molecularly targeted drug administration. Clin Cancer Res 21(20):4586–4596. https://doi.org/10.1158/1078-0432.CCR-15-0584
Jovelet C, Ileana E, Le Deley MC, Motte N, Rosellini S, Romero A, Lefebvre C, Pedrero M, Pata-Merci N, Droin N, Deloger M, Massard C, Hollebecque A, Ferte C, Boichard A, Postel-Vinay S, Ngo-Camus M, De Baere T, Vielh P, Scoazec JY, Vassal G, Eggermont A, Andre F, Soria JC, Lacroix L (2016) Circulating cell-free tumor DNA analysis of 50 genes by next-generation sequencing in the prospective MOSCATO trial. Clin Cancer Res 22(12):2960–2968. https://doi.org/10.1158/1078-0432.CCR-15-2470
Dietz S, Schirmer U, Merce C, von Bubnoff N, Dahl E, Meister M, Muley T, Thomas M, Sultmann H (2016) Low input whole-exome sequencing to determine the representation of the tumor exome in circulating DNA of non-small cell lung cancer patients. PLoS ONE 11(8):e0161012. https://doi.org/10.1371/journal.pone.0161012
Sudmant PH, Rausch T, Gardner EJ, Handsaker RE, Abyzov A, Huddleston J, Zhang Y, Ye K, Jun G, Fritz MH, Konkel MK, Malhotra A, Stutz AM, Shi X, Casale FP, Chen J, Hormozdiari F, Dayama G, Chen K, Malig M, Chaisson MJP, Walter K, Meiers S, Kashin S, Garrison E, Auton A, Lam HYK, Mu XJ, Alkan C, Antaki D, Bae T, Cerveira E, Chines P, Chong Z, Clarke L, Dal E, Ding L, Emery S, Fan X, Gujral M, Kahveci F, Kidd JM, Kong Y, Lameijer EW, McCarthy S, Flicek P, Gibbs RA, Marth G, Mason CE, Menelaou A, Muzny DM, Nelson BJ, Noor A, Parrish NF, Pendleton M, Quitadamo A, Raeder B, Schadt EE, Romanovitch M, Schlattl A, Sebra R, Shabalin AA, Untergasser A, Walker JA, Wang M, Yu F, Zhang C, Zhang J, Zheng-Bradley X, Zhou W, Zichner T, Sebat J, Batzer MA, McCarroll SA, Genomes Project C, Mills RE, Gerstein MB, Bashir A, Stegle O, Devine SE, Lee C, Eichler EE, Korbel JO (2015) An integrated map of structural variation in 2,504 human genomes. Nature 526(7571):75–81. https://doi.org/10.1038/nature15394
Whelan S, Goldman N (2004) Estimating the frequency of events that cause multiple-nucleotide changes. Genetics 167(4):2027–2043. https://doi.org/10.1534/genetics.103.023226
Wyatt AW, Annala M, Aggarwal R, Beja K, Feng F, Youngren J, Foye A, Lloyd P, Nykter M, Beer TM, Alumkal JJ, Thomas GV, Reiter RE, Rettig MB, Evans CP, Gao AC, Chi KN, Small EJ, Gleave ME (2017) Concordance of circulating tumor DNA and matched metastatic tissue biopsy in prostate cancer. J Natl Cancer Inst. https://doi.org/10.1093/jnci/djx118
Shah SP, Roth A, Goya R, Oloumi A, Ha G, Zhao Y, Turashvili G, Ding J, Tse K, Haffari G, Bashashati A, Prentice LM, Khattra J, Burleigh A, Yap D, Bernard V, McPherson A, Shumansky K, Crisan A, Giuliany R, Heravi-Moussavi A, Rosner J, Lai D, Birol I, Varhol R, Tam A, Dhalla N, Zeng T, Ma K, Chan SK, Griffith M, Moradian A, Cheng SW, Morin GB, Watson P, Gelmon K, Chia S, Chin SF, Curtis C, Rueda OM, Pharoah PD, Damaraju S, Mackey J, Hoon K, Harkins T, Tadigotla V, Sigaroudinia M, Gascard P, Tlsty T, Costello JF, Meyer IM, Eaves CJ, Wasserman WW, Jones S, Huntsman D, Hirst M, Caldas C, Marra MA, Aparicio S (2012) The clonal and mutational evolution spectrum of primary triple-negative breast cancers. Nature 486(7403):395–399. https://doi.org/10.1038/nature10933
Garcia-Saenz JA, Ayllon P, Laig M, Acosta-Eyzaguirre D, Garcia-Esquinas M, Montes M, Sanz J, Barquin M, Moreno F, Garcia-Barberan V, Diaz-Rubio E, Caldes T, Romero A (2017) Tumor burden monitoring using cell-free tumor DNA could be limited by tumor heterogeneity in advanced breast cancer and should be evaluated together with radiographic imaging. BMC Cancer 17(1):210. https://doi.org/10.1186/s12885-017-3185-9
Quail MA, Smith M, Coupland P, Otto TD, Harris SR, Connor TR, Bertoni A, Swerdlow HP, Gu Y (2012) A tale of three next generation sequencing platforms: comparison of Ion Torrent, Pacific Biosciences and Illumina MiSeq sequencers. BMC Genomics 13:341. https://doi.org/10.1186/1471-2164-13-341
Hwang S, Kim E, Lee I, Marcotte EM (2015) Systematic comparison of variant calling pipelines using gold standard personal exome variants. Sci Rep-Uk. https://doi.org/10.1038/Srep17875
Kroigard AB, Thomassen M, Laenkholm AV, Kruse TA, Larsen MJ (2016) Evaluation of nine somatic variant callers for detection of somatic mutations in exome and targeted deep sequencing data. PLoS ONE. https://doi.org/10.1371/journal.pone.0151664
Cibulskis K, Lawrence MS, Carter SL, Sivachenko A, Jaffe D, Sougnez C, Gabriel S, Meyerson M, Lander ES, Getz G (2013) Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol 31(3):213–219. https://doi.org/10.1038/nbt.2514
Koboldt DC, Zhang Q, Larson DE, Shen D, McLellan MD, Lin L, Miller CA, Mardis ER, Ding L, Wilson RK (2012) VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res 22(3):568–576. https://doi.org/10.1101/gr.129684.111
Saunders CT, Wong WS, Swamy S, Becq J, Murray LJ, Cheetham RK (2012) Strelka: accurate somatic small-variant calling from sequenced tumor-normal sample pairs. Bioinformatics 28(14):1811–1817. https://doi.org/10.1093/bioinformatics/bts271
Henriquez-Hernandez LA, Murias-Rosales A, Gonzalez-Hernandez A, de Leon AC, Diaz-Chico N, Fernandez-Perez L (2010) Distribution of TYMS, MTHFR, p53 and MDR1 gene polymorphisms in patients with breast cancer treated with neoadjuvant chemotherapy. Cancer Epidemiol 34(5):634–638. https://doi.org/10.1016/j.canep.2010.06.013
Henriquez-Hernandez LA, Perez LF, Hernandez AG, de Leon AC, Diaz-Chico B, Rosales AM (2010) TYMS, MTHFR, p53 and MDR1 gene polymorphisms in breast cancer patients treated with adjuvant therapy. Cancer Epidemiol 34(4):490–493. https://doi.org/10.1016/j.canep.2010.03.004
Acknowledgements
We are grateful to the patients who participated in this study and their family members who supported them. We thank the Norris Cotton Cancer Center Genomics and Pathology Shared Resources for assistance.
Funding
This study was funded by Norris Cotton Cancer Center (Hopeman Foundation Pilot Grant to MDC and TWM), and the NIH (F30CA216966 to KS; R01CA200994 and R01CA211869 to TWM; Dartmouth College Norris Cotton Cancer Center Support Grant P30CA023108).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
All procedures performed involving human subjects were approved by the Dartmouth College Institutional Review Board and were consistent with the 1996 version of the Declaration of Helsinki.
Informed consent
Informed consent was obtained from all individual participants included in the study.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Chamberlin, M.D., Wells, J.D., Shee, K. et al. Plasma DNA as a “liquid biopsy” incompletely complements tumor biopsy for identification of mutations in a case series of four patients with oligometastatic breast cancer. Breast Cancer Res Treat 182, 665–677 (2020). https://doi.org/10.1007/s10549-020-05714-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10549-020-05714-2