Abstract
The construction of a rapid and easy immunofluorescence bioassay for SARS-CoV-2 detection is described. We report for the first time a novel one-pot synthetic approach for simultaneous photoinduced step-growth polymerization of pyrene (Py) and ring-opening polymerization of ε-caprolactone (PCL) to produce a graft fluorescent copolymer PPy-g-PCL that was conjugated to SARS-CoV-2-specific antibodies using EDC/NHS chemistry. The synthesis steps and conjugation products were fully characterized using standard spectral analysis. Next, the PPy-g-PCL was used for the construction of a dot-blot assay which was calibrated for applications to human nasopharyngeal samples. The analytical features of the proposed sensor showed a detection range of 6.03–8.7 LOG viral copy mL−1 (Ct Scores: 8–25), the limit of detection (LOD), and quantification (LOQ) of 1.84 and 6.16 LOG viral copy mL−1, respectively. The repeatability and reproducibility of the platform had a coefficient of variation (CV) ranging between 1.2 and 5.9%. The fluorescence-based dot-blot assay was tested with human samples. Significant differences were observed between the fluorescence intensity of the negative and positive samples, with an overall correct response of 93.33%. The assay demonstrated a high correlation with RT-PCR data. This strategy opens new insights into simplified synthesis procedures of the reporter molecules and their high potential sensing and diagnosis applications.
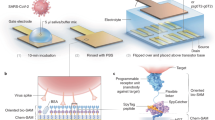
Graphical abstract

Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Optical biosensors are devices that use various materials such as fluorescence, luminescence, and colorimetric molecules offering a myriad of benefits in simplifying the use and lowering the cost of preparation. They are often referred to as the future of point-of-care (POC)-based diagnostics due to the high accuracy and low requirement needed to operate [1]. Their application covers a wide range of targets, including viral detection, substance use, toxins, environmental monitoring, food safety, and many others [2]. Immunoassays are qualitative and quantitative tools for detecting and measuring various targets based on the interaction between an antibody and a target antigen, making them a highly sensitive and selective instrument for diagnosis [3]. Immunoassay approaches have been integrated into many POC tests, such as paper-based assays (lateral flow assays and dot blots). Paper-based immunoassays can be of great economic value due to their simple manufacturing and low cost, which is advantageous in large-scale production and application [4]. Combining these tools with the advantages of fluorescent tags further enhances the potential of paper-based biosensors by increasing their sensitivity. Hence, developing novel combinations of antibodies and fluorescent molecules is essential in biosensor applications.
With continuous advancements in the production of polymers, the development of photochemical synthetic methods has been the subject of extensive research as they afford environmentally benign and sustainable conditions [5]. Conventional synthesis methods utilizing metal catalysts and high temperatures may not be suitable for certain applications [6]. Nowadays, light-activated polymerizations are more favorable in material science due to the inherent benefits offered such as safety, less energy consumption, spatiotemporal control, high efficiency at room temperature, and reduced emissions of volatile organic compounds [7]. We have recently shown that highly conjugated polymers can be prepared by photoinduced step-growth polymerization [8]. Pyrene polymerization is an excellent example of this approach [9] (Fig. 1A).
Homo and copolymers of polylactones, particularly poly(ε-caprolactone) (PCL), are widely used in bio-applications due to their biocompatibility and biodegradability. Various combinations of PCL with other types of polymers imparting biocompatibility to the synthetic polymers were reported [10]. PCL is generally prepared by ring-opening polymerization (ROP) using stannous(II)octoate combined with hydroxyl functional compounds [11]. However, due to metallic contaminations of the polymer products, their bio-applications are quite challenging. Some solutions were proposed to overcome this issue using a phosphazene base [12] or Lewis acids and acid–base organocatalytic systems [13]. We have recently introduced a new photoinduced ROP of ε-CL by using onium salts [14].
As the same active species are involved in both photoinduced step-growth polymerization of pyrene and ROP of ε-CL, it seemed appropriate to conduct both processes simultaneously by using appropriately selected combinations. This study reports for the first time a one-pot in situ preparation of polypyrene-g-poly(ε-caprolactone) (PPy-g-PCL) via simultaneous photoinduced step-growth and ROP polymerization to produce a fluorescent molecule with interesting optical applications.
Since the rise of COVID-19, POC detection is increasingly regarded as a promising method to facilitate home testing and on-site diagnosis [15]. During the 2020–2021 period, the development of reliable methods meeting mass testing needs and reducing costs has attracted attention because of the high testing requirements. The RT-PCR technique, considered a gold-standard technique for SARS-CoV-2 detection, suffers from long reporting time, need for expensive equipment, and trained personnel. Alternately, recent POC-based studies have aimed to design and fabricate straightforward and cheap diagnostic tools capable of identifying target analytes with high sensitivity and specificity [16, 17].
The current study proposes an original fluorescent bioassay setup for SARS-CoV-2 detection. The platform was designed as an optical paper-based dot-blot assay using a fluorescent PPy-g-PCL synthesized with an original approach of a one-pot photoinduced step-growth and ROP polymerization combined with a polyclonal antibody cocktail targeting SARS-CoV-2. The synthesis and bioconjugation steps were fully characterized using various spectral analyses. The biosensor was calibrated and tested with human nasopharyngeal samples to demonstrate its enhanced sensitivity compared with the traditional RT-PCR technique.
Experimental section
Materials
ε-Caprolactone (ε-CL) (Aldrich, 97%) was vacuum distilled over calcium hydride. Pyrene (Py) (Sigma-Aldrich, 98%) was recrystallized from ethanol. Pyrene methanol (PyMe), diphenyliodonium hexafluorophosphate (DPI), succinic anhydride, pyridine, triethylamine were obtained from Sigma-Aldrich (Germany) and used as purchased. Dichloromethane was purified using conventional drying and distillation procedures. Affinity column (protein A) was obtained from Thermo Fisher. Nitrocellulose membranes were ordered from mdi Membrane Technologies (India).
Characterization
Gel permeation chromatography (GPC) measurements were performed on a TOSOH EcoSEC GPC system equipped with an autosampler system, a temperature-controlled pump, a column oven, a refractive index (RI) detector, a purge and degasser unit, and a TSK gel super hZ2000 4.6 mm ID × 15 cm × 2.0 cm column. Tetrahydrofuran (THF) was used as an eluent at a flow rate of 1.0 mL min−1 at 40 °C. A refractive index detector was calibrated with polystyrene standards having narrow molecular weight distributions. SEC (size-exclusion chromatography) data were analyzed using EcoSEC Analysis software. UV − vis spectra were recorded with a Shimadzu UV-1601 double-beam spectrometer equipped with a 50-W halogen lamp and a deuterium lamp which operate between 190 and 1100 nm. All fluorescence spectra were recorded using a PerkinElmer LS55 spectrometer performing between 200 and 900 nm wavelengths with a 10-nm slit width. The 1H NMR spectra of the products were measured at room temperature in Si(CH3)4 using a 500 MHz NMR (Agilent NMR System VNMRS). Differential scanning calorimetry (DSC) was performed on a PerkinElmer Diamond DSC from 30 to 300 °C with a heating rate of 10 °C min−1 under a nitrogen flow. A typical DSC sample was 2.0 − 5.0 mg in a 30-μL aluminum pan. The Fourier transform infrared (FT-IR) spectroscopy measurements were recorded at 16 scans using a PerkinElmer FT-IR.
Polymerization procedures
ε-CL (10 mol L−1), Py (0.25 mol L−1), (PyMe) (0.1 mol L−1), and DPI (0.5 mol L−1) were dissolved in 2.0 mL dry dichloromethane and then placed into a 15-mL Schlenk tube that had been degassed using three freeze pump thaw cycles. The solution was then exposed to light from 12 Philips 7.1-W fluorescent lamps providing light at 350 nm in a photoreactor. The light intensity was ~ 3.0 mW cm−2. After irradiation, the resulting polymer solution was precipitated by pouring it into a tenfold methanol solution. The collected polymer was filtered and dried for 24 h at 50 °C under a vacuum. Gravimetric analysis was used to determine polymers conversions.
Human samples
Human nasopharyngeal swabs (n = 25) and serums (n = 25) were collected at the University Hospital of Ege University, Izmir (Turkey). Sample collection was performed according to standard procedures, and each sample was analyzed with RT-PCR and commercial ELISA tests to determine negative or COVID-19 positive patients (including the English variant). The swab samples were blindly grouped and stocked at – 20 °C till further use, and serum samples were stored similarly till purification steps were initiated. The study on human samples was approved by the Clinical Research Ethics Committee of Ege University under the reference number: 20-8 T/28.
Antibody purification and characterization
Serum samples from infected patients (determined by RT-PCR) were analyzed via commercial ELISA tests to detect and determine COVID-19-specific IgG, IgA, and IgM. Samples with the highest antibody amounts were selected and pooled together to enhance the yield of the antibodies after purification. Pooled samples were subjected to conventional precipitation, and the precipitate was dialyzed overnight before being applied over a protein A/G affinity column. The eluted antibodies were concentrated, aliquoted, and stored at − 20 °C till further use [18].
The antibody concentration was determined using UV measurements (280 nm). SDS-PAGE analysis was also performed to assess the purity of the antibodies. The specificity of the antibodies was confirmed through two different approaches: SARS-CoV-2-specific ELISA kits for (IgG, IgA, and IgM) and electrochemical-based analysis using a screen-printed electrode coupled with magnetic nanoparticles-conjugated commercial antigens (protein S1 and S2) [18]. A detailed protocol of the purification and characterization can be seen in the Supplementary Information.
Application of the PPy-g-PCL in paper-based biosensing of COVID-19
PPy-g-PCL conjugation
The conjugation of the COVID-19 antibodies to PPy-g-PCL was performed in a similar way to our previous procedure of antibody-polymersome conjugation using EDC/NHS chemistry [19]. The first step consisted of introducing a carboxylic group over the hydroxyl end group of PPy-g-PCL using succinic anhydride with pyridine/triethylamine as the catalyst. In brief, PPy-g-PCL (1.0 eq.), succinic anhydride (2 eq.), and pyridine/triethylamine (1:1) (1.0 eq.) were dispersed in 10 volumes of 1,4-dioxane and left to react for 24 h at room temperature. Later, the carboxylated PPy-g-PCL was sedimented through repeated treatments with hexane/ether (1:1) three times.
The obtained powder (10 mg) was reacted with EDC/NHS in MES buffer according to the previously described approach [20]. Later, the SARS-CoV-19 antibodies (1.0 mg/mL in PBS) were added over the functionalized PPy-g-PCL and stirred at room temperature for 24 h under argon gas. The solution was dialyzed overnight, and antibody-PPy-g-PCL was stored at + 4 °C till further use. The different steps of the bioconjugation were characterized using fluorimetry (Varian Cary eclipse, Perkin Elmer Ltd., England) and FT-IR analyses (Perkin Elmer Spectrum Two). The fluorescence excitation measurements were performed using an emission wavelength of 390 nm.
Paper-based dot-blot assay for virus detection
The dot-blot assay for the COVID-19 sensing using PPy-g-PCL conjugated antibodies was performed in a manner similar to the procedure previously reported using antibody-conjugated polymersomes [19]. Briefly, 1.5 mg mL−1 of antibody (2.0 μL) was spotted over a nitrocellulose membrane and left at room temperature overnight. The spots received 2.0 μL of BSA (1.0%) to block non-specific sites for 30 min at 37 °C, followed by three washes using PBS. After drying, the different nasopharyngeal samples (positive and negatives) were added over the prepared spots (2.0 μL) and incubated for 30 min at 37 °C. After another round of washing and drying, 2.0 μL of PPy-g-PCL conjugated antibodies solution (1:40 dilution in ethanol 20%) was added, and the membranes were incubated for 30 min at 37 °C. The membranes were rinsed with PBS (2.0 μL) and taken for smartphone-assisted image analysis. A blank spot receiving no human sample was also prepared to determine the background fluorescence.
Fluorescence analysis using smartphone-assisted imaging
Membranes were photographed before and after being put under a UV lamp to reveal fluorescence using a Xiaomi smartphone (Redmi Note 9 Pro). The pictures were taken from a distance of 20 cm and transferred for analysis using Image J software (NIH, Maryland, USA). The selection of the fluorescent regions was performed using the ellipse tool, and great care was taken to avoid the ring forming at the periphery of the spots. The images were then measured for signal intensity and surface area of the fluorescence using the Analyze option [19]. The data of the blank spots were subtracted from all others to eliminate the background interference, and the obtained values were divided by the surface area to get a corrected fluorescence intensity. A detailed demonstration of the image-based fluorescence analysis is provided (Fig. S1).
Calibration and analytical performance
The calibration of the dot-blot assay was performed using viral load (copy mL−1) determined from the RT-PCR data (106 to 1011 viral copy mL−1 equivalent to 8–25 Ct scores) according to the previously described report [21]. The fluorescence intensity was determined, and data was plotted in function of viral load or LOG10 of viral copy number to determine the analytical performance of the biosensor, including the detection range, limit of detection (LOD), and quantification (LOQ). The sensitivity and selectivity of the biosensor were analyzed through a correlation study between the current platform and the data generated from the RT-PCR analysis (Ct scores). Additionally, the repeatability and reproducibility of the tests were investigated by taking samples from each group and testing over the same or different batches of the dot-blot assay (n = 10). The data was plotted, and the overall response and coefficient of variation were calculated.
Statistical analysis
The analyses were performed at least three times, and the results are shown as mean ± SD. The statistical differences were analyzed standard t-test, and p values lower than 0.05 were considered significant.
Results and discussion
The current study addresses the development of a fluorescent paper-based dot-blot immunoassay to detect SARS-CoV-2 antigens in human nasopharyngeal samples (Fig. 2). Since the pandemic’s start, there have been continuous efforts in proposing efficient sensors for fast and reliable screening. Yet, RT-PCR and paper-based assays remain the major diagnostic tools employed. Of interest to the current study, paper-based assays are broadly spread worldwide for both antigen and antibody detection due to their fast turnaround, cost-effectiveness, and user-friendliness. Colorimetric immunoassays especially make use of gold nanoparticles as a color reporter. Albeit the proven performance of these molecules, there are some disadvantages related to stability and color bleaching artifacts. From here, enhancing the assay format has seen various efforts, including the affinity and recognition materials (antibodies, aptamers, enzymes, genetic materials, etc.), assay materials (nitrocellulose, polydimethylsiloxane, etc.), and reporting optical materials (dyes, nanoparticles, polymersomes, fluorescent molecules, etc.). The use of colorimetric materials such as dye-loaded polymersome has been shown to perform well compared with gold nanoparticles and was shown to have a high correlation with PCR data [19]. The subject of sensitivity to detect small amounts of targets remained an open door for development, and it has been shown that the employment of fluorescent molecules can enhance the optical capabilities of paper-based assays [22]. Taking advantage of these fluorescent molecules, the current study reports for the first time a novel approach for the synthesis of a fluorescent polypyrene-g-poly(ε-caprolactone) (PPy-g-PCL) prepared by simultaneous photoinduced step-growth and ROP processes for paper-based SARS-CoV-2 sensing application.
Fluorescent PPy-g-PCL synthesis
Chain polymerizations involving free radicals, cationic, and anionic routes can be accomplished by photochemical means in the broad wavelength range covering the UV [23], visible [24], and near-IR [25] regions of the electromagnetic spectrum. In these processes, upon irradiation, the excited conjugated monomer forms an exciplex that undergoes a single electron transfer reaction with iodonium salt. Thus, the formed monomer radical cation releases protons, resulting in monomer radicals that couple with each other. Photocatalytic cycles of electron transfer, proton release, and radical coupling steps essentially yield desired highly conjugated polymers. The latest reports from our laboratory have demonstrated that this photochemical approach can successfully be applied for the synthesis of polythiophenes [26], polycarbazoles [27], and polypyrenes [9].
PCL is a widely used polymer in biomedical applications due to its various advantages conferring biocompatibility features to many polymers combined with it [10]. While PCL is synthesized through ROP, its synthesis shares many active species with the photoinduced step-growth polymerization of pyrene. As such, the design of a single step, one-pot simultaneous synthesis to produce a PPy-g-PCL appeared to be feasible to avoid metallic contaminations during the preparation steps.
We first studied simultaneous step-growth polymerization of pyrene and ROP of CL by photoinduced electron transfer (PET) to yield the desired fluorescent base polymer (Fig. 1B). In this process, the pyrene acted as a light-absorbing sensitizer and monomer. Pyrene methanol was deliberately selected to act as a ROP initiator and provide attachment for PCL segments to pyrene repeating units. The reaction showed that the yield and molecular weight increased with the irradiation time (Table 1). The relatively high dispersity observed reflects the characteristic nature of step-growth polymerization.
Because of its improved stability and flexibility, the PCL segment was deliberately selected as a biocompatible segment [28]. Moreover, its photochemical synthetic approach is highly compatible with the step-growth polymerization of pyrene [9]. The successful synthesis and structure of the polymer were confirmed by spectral analyses, including UV–Vis, fluorescence, FT-IR, and 1H NMR (Fig. S2), and DSC investigations (Fig. S3). The analysis results and related information can be seen in the Supplementary Information.
PPy-g-PCL-antibody conjugation
The first step consisted of purifying highly sensitive antibodies from human samples through standard affinity-based methods. The produced antibodies were carefully characterized by various techniques such as ELISA, SDS-PAGE, LC-Q-TOF–MS/MS, and electrochemical analysis to validate the purity and functionality of the proteins [3, 18]. The purified antibodies were conjugated with the synthesized PPy-g-PCL using succinic anhydride and EDC/NHS chemistries. Successful bioconjugation was confirmed by fluorescence and FT-IR spectral analyses (Fig. S4). Detailed assessment of the characterization data can be seen in the Supplementary Information.
To further characterize the interaction of the PPy-g-PCL-antibody conjugate with the human samples over the dot-blot system, scanning electron microscopy (SEM) and fluorescence microscopy investigations were performed. The SEM observation demonstrated that positive and negative samples provide distinct changes to the physical organization and surface of the polymer-antibody conjugates (Fig. 3). The addition of human samples induces aggregation of the PPy-g-PCL-antibody conjugates that can be attributed to the antigen’s recognition by the antibodies. The negative sample showing some accumulation is expected due to the possibility of non-specific interactions or physical entrapment. On the other hand, the positive sample addition shows burgeoning structures due to the surrounding of the polymer-antibody conjugates around the antigens (Fig. 3 top row). Fluorescence microscopy further confirms the SEM observations with the positive samples inducing a significant aggregation of the PPy-g-PCL-antibody conjugates around the antigen targets, resulting in enhanced fluorescent regions (Fig. 3 bottom row).
SARS-CoV-2 biosensor construction and analytical evaluation
Following the successful synthesis of PPy-g-PCL and its antibody conjugate, a fluorescence paper-based dot-blot assay for detecting SARS-CoV-2 was developed. The system is based on a sandwich-type of immunosensing with antibodies immobilized over the nitrocellulose membrane followed by the sample for detection and then another addition of an antibody-PPy-g-PCL conjugate for revelation (Fig. 2). The biological application of the fluorescent bioconjugate was performed over human samples applied over a dot-blot assay by the technique reported in our earlier work [19] with some modifications (Fig. 2). The membranes were then photographed under UV light to develop the fluorescence related to the presence of the target. Under normal light, the proposed detection platform showed circular spots with light brownish colors indicating no differences between positive and negative samples. However, the fluorescence induction provides a distinct difference between negative and positive samples that can be seen from the color intensity of the dot-blot surface.
The calibration procedure of the dot-blot assay was performed using human nasopharyngeal samples of known viral load. The estimation of viral copy numbers was calculated from RT-PCR calibration data of the analysis bodies of the Ege University Hospital following standard procedures [29]. The analysis of increasing viral loads induced improved fluorescence intensities (Fig. 4A, B). The linear detection range of the assay was estimated between 6.03 and 8.7 LOG viral copy mL−1 (y = 13.661x + 49.75; R2 = 0.9915). The limit of detection (LOD) and limit of quantification (LOQ) were estimated using the LOD = 3 × SD/m, and LOQ = 10 × SD/m equations (with SD: standard deviation of the intercept and m: the slope). The proposed biosensor showed a great sensitivity with LOD of 1.84 and LOQ of 6.16 LOG viral copy mL−1. Furthermore, the testing time of sample and conjugate application and the waiting time for color development were estimated at 15–20 min.
Analytical performance of the proposed PPy-g-PCL-based dot-blot assay for detecting SARS-CoV-2. Calibration analysis of the proposed fluorescent biosensor expressed in A viral load (viral copy mL−1) and B LOG10 viral load. The fluorescence was induced using a lamp (350–390 nm). C Corrected fluorescence intensities measured for the different nasopharyngeal samples and representative visual and fluorescent observations of the spots (insets). D Correlation of the PPy-g-PCL-based dot-blot sensor’s data with the RT-PCR Ct scores. E, F Repeatability and reproducibility of the assay and G the response performance of the biosensor (overall or by subgroups). CV is the coefficient of variation between the repetitive measurements expressed in percentages. ***p < 0.001 vs. negative group. ns, non-significant
Many biosensor platforms have been proposed for detecting SARS-CoV-2 in the literature ranging from traditional approaches such as ELISA to other highly sensitive tools like molecularly imprinted polymer sensors. These sensors have various targets, detection ranges, and sensitivity levels that contribute to their advantage and disadvantage and the reliability of their implementation. We have collected some of the recent and best-performing sensors using polymeric structures reported in the literature to compare with the analytical performance of the current fluorescent dot-blot assay (Table 2). Including this work, some polymer structures are used as labels in colorimetric assays [3, 30]. Other sensors make use of polymers as immobilization matrices [31,32,33] or biorecognition surfaces [34, 35]. There is a limited number of research studies on the development of SARS-CoV-2 diagnostic tools based on polymers and co-polymeric structures. Hence, the current work provides a substantial addition to the subject, especially with the novel synthetic method to produce fluorescent molecules.
Application of PPy-g-PCL in SARS-CoV-2 sensing
The validation of the proposed biosensor was performed over infected human serums. The nasopharyngeal samples were pre-tested with RT-PCR to identify positive and negative samples. The same samples were then tested over the fluorescent dot-blot assay. The analysis of the images for quantifying the fluorescence levels showed a significant difference between the negative and positive samples (p < 0.001). Interestingly, the proposed platform showed a similar detection capacity for both SARS-CoV-2 positive and UK variant samples (Fig. 4C). The overall correlation of the current results with the PCR-obtained Ct scores demonstrated a value of − 0.92, suggesting a high correlation towards the number of virus particles in the samples (Fig. 4D). This linearity highlights the sensitivity of the PPy-g-PCL-based dot-blot assays that provide results similar to the RT-PCR.
The repeatability of results is an essential criterion for developing any biosensor. It provides information on the applicability of the platform at larger cohorts and gives an idea of the reliability of the test under variations that can occur during measurements. The current SARS-CoV-2 dot-blot sensor showed a high level of repeatability with a coefficient of variation (CV) ranging between 4.4 and 5.9% (Fig. 4E). Similarly, the reproducibility was also explored through testing samples over various batches of freshly prepared dot-blot assays. Indeed, the results showed similar results to the repeatability analysis with a CV% ranging between 1.2 and 2.5% (Fig. 4F). This is of high importance because the CV% in biosensor development is recommended to be below 10% to be considered reliable [36].
The overall response of the proposed sensor was estimated as 93.3% correct diagnosis compared to RT-PCR (Fig. 4G), with 3.3% false diagnosis coming from the group of the UK variant. The separation of each subgroup demonstrated that the response of the proposed dot-blot assay had 100% correct diagnosis of the negative and positive samples (n = 20). In contrast, the UK variant group had 2 of the ten samples that did not respond correctly. The biosensor had no false positive and 6.6% false negative.
Limitations and perspectives
Development in diagnostic platforms generally aims for fast, cheap, user-friendly tools, and, most importantly, reliable results. During the current COVID-19 pandemic, significant advances in the field of portable diagnostic and point-of-care (POC) devices have been achieved. The applications and advantages of optical paper-based assays are well known, yet, they have their share of limitations as many other techniques employed. Generally, sensitivity is an essential issue for paper-based assays due to the need for high concentrations of the target molecule to produce a colorimetric signal visible by the naked eye [37]. The proposed assay might suffer from sensitivity with low viral load samples (Ct scores > 25). Due to the unavailability of such samples, proper testing was not performed to check the sensitivity of the current assay at low viral loads. It would be interesting to enhance the sample numbers representing a wider Ct score range to obtain fine-tuned data on the capabilities of the current dot-blot assay. Albeit missing, the correlation of the sensor data with the RT-PCR scores demonstrated a high correlation suggesting that the sensor will perform well with any viral load.
Another issue with the rapid antigen tests resides in the increasing number of variants (specifically in the case of SARS-CoV-2), which is reflected by the continuous changes in the target structures, thus producing increasing numbers of false-negative diagnoses. Indeed, many reports and the World Health Organization (WHO) suggested the targeting of nucleocapsid (N) proteins or a combination of N and spike (S) proteins [38]. There are various solutions proposed to overcome the variants issue, such as using protein target regions that are less susceptible for mutations, or as in the case of the current work, the use of a combination of monoclonal or polyclonal antibody cocktails to increase the sensitivity of the biosensor [39].
The use of fluorescent molecules as reporting agents proved to enhance the sensitivity of the paper-based biosensors [22]. However, such an approach raises a challenge in portability and POC applications due to the need for fluorescence-inducing apparatuses for signal induction. Thankfully, the current technological progress has provided many solutions for this issue. Many research groups have been contributing to the subject of PCT technologies by developing optomechanical devices that can help image acquisition with different variables for biomedical applications [40]. From this, it can be interesting to create optomechanical tools with integrated UV light for facilitating on-site imaging and analysis.
Conclusion
The combination of fluorescent conjugates and biocompatible polymers is at the verge of a new era with important implications for diverse fields and applications, ranging from biosensors, immunoassays, bio-surfaces to synthetic photochemistry. Simultaneous photoinduced step-growth polymerization and ROP were proposed and achieved to prepare fluorescent graft copolymers possessing both conjugated PPy and PCL building blocks. Our work shows that the reactive species formed from the PET reactions between excited pyrene and iodonium salt play a key role in inducing both polymerizations concomitantly. After bio-functionalization of the fluorescent graft copolymer with SARS-CoV-2 recognizing antibodies, it was successfully adapted into a paper-based bioassay system as a labeling agent. The analytical features demonstrated high sensitivity and correlation with RT-PCR. These data are valuable for future potential usage of the PPy-g-PCL graft copolymer as an essential component of fluorescence-based diagnostic kits and other biosensors.
Data availability
All data are included in the current manuscript and Supplementary Information. Raw data are available upon request.
Code availability
Not applicable.
References
Saylan Y, Erdem O, Unal S, Denizli A (2019) An alternative medical diagnosis method: biosensors for virus detection. Biosensors (Basel) 9(2):65. https://doi.org/10.3390/bios9020065
Aydindogan E, Balaban S, Evran S, Coskunol H, Timur S (2019) A bottom-up approach for developing aptasensors for abused drugs: biosensors in forensics. Biosensors (Basel) 9(4):118. https://doi.org/10.3390/bios9040118
Ghorbanizamani F, Tok K, Moulahoum H, Harmanci D, Hanoglu SB, Durmus C, Zihnioglu F, Evran S, Cicek C, Sertoz R, Arda B, Goksel T, Turhan K, Timur S (2021) Dye-loaded polymersome-based lateral flow assay: rational design of a COVID-19 testing platform by repurposing SARS-CoV-2 antibody cocktail and antigens obtained from positive human samples. ACS Sens 6(8):2988–2997. https://doi.org/10.1021/acssensors.1c00854
Huang Y, Xu T, Wang W, Wen Y, Li K, Qian L, Zhang X, Liu G (2019) Lateral flow biosensors based on the use of micro- and nanomaterials: a review on recent developments. Mikrochim Acta 187(1):70. https://doi.org/10.1007/s00604-019-3822-x
Xiao P, Zhang J, Dumur F, Tehfe MA, Morlet-Savary F, Graff B, Gigmes D, Fouassier JP, Lalevee J (2015) Visible light sensitive photoinitiating systems: Recent progress in cationic and radical photopolymerization reactions under soft conditions. Prog Polym Sci 41:32–66. https://doi.org/10.1016/j.progpolymsci.2014.09.001
Hillmyer MA, Tolman WB (2014) Aliphatic polyester block polymers: renewable, degradable, and sustainable. Acc Chem Res 47(8):2390–2396. https://doi.org/10.1021/ar500121d
Fouassier J-P, Lalevée J (2012) Photoinitiators for polymer synthesis: scope, reactivity, and efficiency. Wiley, Weinheim
Yilmaz G, Yagci Y (2020) Light-induced step-growth polymerization. Prog Polym Sci 100:101178. https://doi.org/10.1016/j.progpolymsci.2019.101178
Celiker T, Kaya K, Koyuncu S, Yagci Y (2020) Polypyrenes by photoinduced step-growth polymerization. Macromolecules 53(14):5787–5794. https://doi.org/10.1021/acs.macromol.0c00694
Degirmenci M, Izgin O, Yagci Y (2004) Synthesis and characterization of cyclohexene oxide functional poly (ε-caprolactone) macromonomers and their use in photoinitiated cationic homo-and copolymerization. J Polym Sci Part A: Polym Chem 42(13):3365–3372
Kowalski A, Duda A, Penczek S (2000) Kinetics and mechanism of cyclic esters polymerization initiated with tin(II) octoate. 3. Polymerization of L,L-dilactide. Macromolecules 33(20):7359–7370. https://doi.org/10.1021/ma000125o
Ladelta V, Kim JD, Bilalis P, Gnanou Y, Hadjichristidis N (2018) Block copolymers of macrolactones/small lactones by a “catalyst-switch” organocatalytic strategy. Thermal Properties and Phase Behavior. Macromolecules 51(7):2428–2436
Arslan M, Kiskan B, Yagci Y (2018) Ring-opening polymerization of 1,3-benzoxazines via borane catalyst. Polymers (Basel) 10(3):239. https://doi.org/10.3390/polym10030239
Bener S, Yilmaz G, Yagci Y (2021) Directly and indirectly acting photoinitiating systems for ring-opening polymerization of ϵ-caprolactone. ChemPhotoChem 5(12):1089–1093. https://doi.org/10.1002/cptc.202100118
Beduk T, Beduk D, de Oliveira Filho JI, Zihnioglu F, Cicek C, Sertoz R, Arda B, Goksel T, Turhan K, Salama KN, Timur S (2021) Rapid point-of-care COVID-19 diagnosis with a gold-nanoarchitecture-assisted laser-scribed graphene biosensor. Anal Chem 93(24):8585–8594. https://doi.org/10.1021/acs.analchem.1c01444
Idili A, Parolo C, Alvarez-Diduk R, Merkoci A (2021) Rapid and efficient detection of the SARS-CoV-2 spike protein using an electrochemical aptamer-based sensor. ACS Sens 6(8):3093–3101. https://doi.org/10.1021/acssensors.1c01222
Jiang N, Tansukawat ND, Gonzalez-Macia L, Ates HC, Dincer C, Guder F, Tasoglu S, Yetisen AK (2021) Low-cost optical assays for point-of-care diagnosis in resource-limited settings. ACS Sens 6(6):2108–2124. https://doi.org/10.1021/acssensors.1c00669
Tok K, Moulahoum H, Ghorbanizamani F, Harmanci D, BalabanHanoglu S, Durmus C, Evran S, Cicek C, Sertoz R, Arda B, Goksel T, Turhan K, Timur S, Zihnioglu F (2021) Simple workflow to repurpose SARS-CoV-2 swab/serum samples for the isolation of cost-effective antibody/antigens for proteotyping applications and diagnosis. Anal Bioanal Chem 413(29):7251–7263. https://doi.org/10.1007/s00216-021-03654-4
Ghorbanizamani F, Moulahoum H, Zihnioglu F, Evran S, Cicek C, Sertoz R, Arda B, Goksel T, Turhan K, Timur S (2021) Quantitative paper-based dot blot assay for spike protein detection using fuchsine dye-loaded polymersomes. Biosens Bioelectron 192:113484. https://doi.org/10.1016/j.bios.2021.113484
Dieu LH, Wu D, Palivan CG, Balasubramanian V, Huwyler J (2014) Polymersomes conjugated to 83–14 monoclonal antibodies: in vitro targeting of brain capillary endothelial cells. Eur J Pharm Biopharm 88(2):316–324. https://doi.org/10.1016/j.ejpb.2014.05.021
Berger A, Nsoga MTN, Perez-Rodriguez FJ, Aad YA, Sattonnet-Roche P, Gayet-Ageron A, Jaksic C, Torriani G, Boehm E, Kronig I, Sacks JA, de Vos M, Bausch FJ, Chappuis F, Renzoni A, Kaiser L, Schibler M, Eckerle I (2021) Diagnostic accuracy of two commercial SARS-CoV-2 antigen-detecting rapid tests at the point of care in community-based testing centers. PLoS ONE 16(3):e0248921. https://doi.org/10.1371/journal.pone.0248921
Moulahoum H, Ghorbanizamani F, Timur S (2021) Paper-based lateral flow assay using rhodamine B-loaded polymersomes for the colorimetric determination of synthetic cannabinoids in saliva. Mikrochim Acta 188(11):402. https://doi.org/10.1007/s00604-021-05062-y
Chen H, Noirbent G, Liu SH, Brunel D, Graff B, Gigmes D, Zhang YJ, Sun K, Morlet-Savary F, Xiao P, Dumur F, Lalevee J (2021) Bis-chalcone derivatives derived from natural products as near-UV/visible light sensitive photoinitiators for 3D/4D printing. Mater Chem Front 5(2):901–916. https://doi.org/10.1039/d0qm00755b
Kiliclar HC, Gencosman E, Yagci Y (2021) Visible light induced conventional step-growth and chain-growth condensation polymerizations by electrophilic aromatic substitution. Macromol Rapid Commun 43:e2100584. https://doi.org/10.1002/marc.202100584
Pang Y, Shiraishi A, Keil D, Popov S, Strehmel V, Jiao H, Gutmann JS, Zou Y, Strehmel B (2021) NIR-sensitized cationic and hybrid radical/cationic polymerization and crosslinking. Angew Chem Int Ed Engl 60(3):1465–1473. https://doi.org/10.1002/anie.202010746
Celiker T, Isci R, Kaya K, Ozturk T, Yagci Y (2020) Photoinduced step-growth polymerization of thieno [3, 4-b] thiophene derivatives. The substitution effect on the reactivity and electrochemical properties. J Polym Sci 58(17):2327–2334
Sari E, Yilmaz G, Koyuncu S, Yagci Y (2018) Photoinduced step-growth polymerization of N-ethylcarbazole. J Am Chem Soc 140(40):12728–12731. https://doi.org/10.1021/jacs.8b08668
Day M, Cooney JD, Shaw K, Watts J (1998) Thermal analysis of some environmentally degradable polymers. J Therm Anal Calorim 52(2):261–274. https://doi.org/10.1023/a:1010195105547
Han MS, Byun JH, Cho Y, Rim JH (2021) RT-PCR for SARS-CoV-2: quantitative versus qualitative. Lancet Infect Dis 21(2):165. https://doi.org/10.1016/S1473-3099(20)30424-2
Zhu X, Wang X, Han L, Chen T, Wang L, Li H, Li S, He L, Fu X, Chen S, Xing M, Chen H, Wang Y (2020) Multiplex reverse transcription loop-mediated isothermal amplification combined with nanoparticle-based lateral flow biosensor for the diagnosis of COVID-19. Biosens Bioelectron 166:112437. https://doi.org/10.1016/j.bios.2020.112437
Ditte K, Le Nguyen TA, Ditzer O, Sandoval Bojorquez DI, Chae S, Bachmann M, Baraban L, Lissel F (2021) Rapid detection of SARS-CoV-2 antigens and antibodies using OFET biosensors based on a soft and stretchable semiconducting polymer. ACS Biomater Sci Eng. https://doi.org/10.1021/acsbiomaterials.1c00727
Nabil A, Yoshihara E, Hironaka K, Hassan AA, Shiha G, Ebara M (2021) Temperature responsive smart polymer for enabling affinity enrichment of current coronavirus (SARS-CoV-2) to improve its diagnostic sensitivity. Comput Struct Biotechnol J 19:3609–3617. https://doi.org/10.1016/j.csbj.2021.06.016
Avelino K, Dos Santos GS, Frias IAM, Silva-Junior AG, Pereira MC, Pitta MGR, de Araujo BC, Errachid A, Oliveira MDL, Andrade CAS (2021) Nanostructured sensor platform based on organic polymer conjugated to metallic nanoparticle for the impedimetric detection of SARS-CoV-2 at various stages of viral infection. J Pharm Biomed Anal 206:114392. https://doi.org/10.1016/j.jpba.2021.114392
Hussein HA, Kandeil A, Gomaa M, Mohamed El Nashar R, El-Sherbiny IM, Hassan RYA (2021) SARS-CoV-2-impedimetric biosensor: virus-imprinted chips for early and rapid diagnosis. ACS Sens 6(11):4098–4107. https://doi.org/10.1021/acssensors.1c01614
Raziq A, Kidakova A, Boroznjak R, Reut J, Opik A, Syritski V (2021) Development of a portable MIP-based electrochemical sensor for detection of SARS-CoV-2 antigen. Biosens Bioelectron 178:113029. https://doi.org/10.1016/j.bios.2021.113029
Chen LC, Wang E, Tai CS, Chiu YC, Li CW, Lin YR, Lee TH, Huang CW, Chen JC, Chen WL (2020) Improving the reproducibility, accuracy, and stability of an electrochemical biosensor platform for point-of-care use. Biosens Bioelectron 155:112111. https://doi.org/10.1016/j.bios.2020.112111
Carrell C, Kava A, Nguyen M, Menger R, Munshi Z, Call Z, Nussbaum M, Henry C (2019) Beyond the lateral flow assay: a review of paper-based microfluidics. Microelectron Eng 206:45–54. https://doi.org/10.1016/j.mee.2018.12.002
Marien J, Ceulemans A, Michiels J, Heyndrickx L, Kerkhof K, Foque N, Widdowson MA, Mortgat L, Duysburgh E, Desombere I, Jansens H, Van Esbroeck M, Arien KK (2021) Evaluating SARS-CoV-2 spike and nucleocapsid proteins as targets for antibody detection in severe and mild COVID-19 cases using a Luminex bead-based assay. J Virol Methods 288:114025. https://doi.org/10.1016/j.jviromet.2020.114025
Chen RE, Zhang X, Case JB, Winkler ES, Liu Y, VanBlargan LA, Liu J, Errico JM, Xie X, Suryadevara N, Gilchuk P, Zost SJ, Tahan S, Droit L, Turner JS, Kim W, Schmitz AJ, Thapa M, Wang D, Boon ACM, Presti RM, O’Halloran JA, Kim AHJ, Deepak P, Pinto D, Fremont DH, Crowe JE Jr, Corti D, Virgin HW, Ellebedy AH, Shi PY, Diamond MS (2021) Resistance of SARS-CoV-2 variants to neutralization by monoclonal and serum-derived polyclonal antibodies. Nat Med 27(4):717–726. https://doi.org/10.1038/s41591-021-01294-w
Brunauer A, Ates HC, Dincer C, Früh SM (2020) Integrated paper-based sensing devices for diagnostic applications. In: Merkoçi A (ed) Paper Based Sensors, vol 89. Comprehensive Analytical Chemistry. Elsevier, pp 397-450. https://doi.org/10.1016/bs.coac.2020.03.003
Funding
The authors would like to acknowledge the financial support from The Scientific and Technological Research Council of Turkey (Project Number: 120C121), Istanbul Technical University Research Fund (Project Number: TUB-2021–43195), and Ege University Research Foundation (project number: TOA-2020–21862). One of the authors (T.C.) would like to thank the Council of Higher Education of Turkey for 100/2000 CoHE Doctoral Scholarship and the Scientific and Technological Research Council of Turkey (TUBITAK) for financial support.
Author information
Authors and Affiliations
Contributions
All the authors contributed significantly to the manuscript and contributed to the redaction. All authors approved the final version of the manuscript. T.C., F.G., H.M., S.T., and Y.Y.: designed the study; T.C., F.G., H.M., and K.T.: performed the experiments and generated the data; C.C., R.S., A.B., T.G., and K.T.: helped in sample collection, analysis, instrumentation, and treatment of experimental data; T.C., E.G.C., and H.M.: wrote the manuscript; E.G.C., F.Z., S.T., and Y.Y.: coordinated, supervised, guided the experimental activities, and revised and gave suggestions during the writing of the manuscript.
Corresponding authors
Ethics declarations
Ethics approval
The study on human samples was approved by the Clinical Research Ethics Committee of Ege University under the reference number 20-8 T/28.
Consent to participate
Not applicable.
Consent for publication
All authors approved the final version of the manuscript.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
604_2022_5244_MOESM1_ESM.docx
Supplementary file1 (DOCX 3407 KB) The online version contains supplementary material available at ImageJ image analysis of the spot fluorescence intensity (Fig. S1). Spectral analysis of the successful synthesis of PPy-g-PCL (Fig. S2). DSC trace of PPy-g-PCL polymer (Fig. S3). Fluorescence and FT-IR spectra of the prepared antibody-PPy-g-PCL conjugate (Fig. S4).
Rights and permissions
About this article
Cite this article
Celiker, T., Ghorbanizamani, F., Moulahoum, H. et al. Fluorescent bioassay for SARS-CoV-2 detection using polypyrene-g-poly(ε-caprolactone) prepared by simultaneous photoinduced step-growth and ring-opening polymerizations. Microchim Acta 189, 202 (2022). https://doi.org/10.1007/s00604-022-05244-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00604-022-05244-2