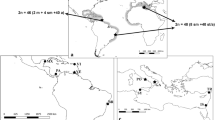
Abstract
Cardinalfishes are a diverse family of small fish found in reef habitats. Some species exhibit bioluminescence and unusual characteristics of buccal egg incubation. Cytogenetic data on the family are confined to Pacific species but reveal remarkable karyotype diversity, as highlighted by low diploid values (2n = 34–46), which likely resulted from centric fusions. Therefore, chromosomal investigations, including samples from different marine regions and with a broader phylogenetic range, are required to elucidate the karyotype history of this group. In this study, we analyzed species from the Atlantic Ocean (Apogon americanus and Phaeoptyx pigmentaria) and the Indo-Pacific region (Sphaeramia nematoptera and Pterapogon kauderni) using conventional (Giemsa staining, Ag-NORs, and C-banding) and molecular (in situ mapping of rDNAs, retrotransposons, and microsatellites) cytogenetic methods. A noticeable karyotype reduction (2n = 46 in S. nematoptera and Pte. kauderni, 2n = 38 in P. pigmentaria, and 2n = 36 in A. americanus) was detected, as well as a decrease in C-positive heterochromatin content (limited to telomeric and centromeric areas). In Indo-Pacific Ocean species, 18S rDNA and 5S rDNA are situated on distinct chromosomes, whereas in Atlantic Ocean species, they are syntenic. Interstitial telomeric sequences were found in S. nematoptera, A. americanus, and P. pigmentaria, indicating that in tandem fusions played a role in the chromosomal decrease in this group. Cardinal fish karyotype variability is remarkable, given the conservative diploid number that characterizes other Percomorpha species. It is probable that biological traits, such as buccal incubation and a brief pelagic larval stage, influence their genetic structure and the rapid rate of chromosomal differentiation.
Graphical abstract





Similar content being viewed by others
References
Allen G. Threatened fishes of the world: Pterapogon kauderni Koumans, 1933 (Apogonidae). Environ Biol Fishes. 2000;57:142. https://doi.org/10.1023/A:1007639909422.
Alvarez MC, Otis J, Amores A, Guise K. Short-term cell culture technique for obtaining chromosomes in marine and freshwater fish. J Fish Biol. 1991;39:817–24. https://doi.org/10.1111/j.1095-8649.1991.tb04411.x.
Ao J, Mu Y, Xiang LX, et al. Genome sequencing of the perciform fish Larimichthys crocea provides insights into molecular and genetic mechanisms of stress adaptation. PLoS Genet. 2015;11: e1005118. https://doi.org/10.1371/journal.pgen.1005118.
Araújo WC, Martínez PA, Molina WF. Mapping of ribosomal DNA by FISH, EcoRI digestion and replication bands in the cardinalfish Apogon americanus (Perciformes). Cytologia. 2010;75:109–17.
Bacon CD, Silvestro D, Jaramillo C, Smith BT, Chakrabarty P, Antonelli A. Biological evidence supports an early and complex emergence of the isthmus of Panama. PNAS. 2015;112:6110–5. https://doi.org/10.1073/pnas.1423853112.
Barnett A, Bellwood DR. Sexual dimorphism in the buccal cavity of paternal mouthbrooding cardinalfishes (Pisces: Apogonidae). Mar Biol. 2005;148:205–12. https://doi.org/10.1007/s00227-005-0052-z.
Bernardi G, Vagell A. Population structure in Banggai cardinalfish, Pterapogon kauderni, a coral reef species lacking a pelagic larval phase. Mar Biol. 2004;145:803–10. https://doi.org/10.1007/s00227-004-1355-1.
Blumer LS. A bibliography and categorization of bony fishes exhibiting parental care. Zool J Linn Soc. 1982;75:1–22. https://doi.org/10.1111/j.1096-3642.1982.tb01939.x.
Bradbury IR, Laurel B, Snelgrove PVR, Bentzen P, Campana SE. Global patterns in marine dispersal estimates: the influence of geography, taxonomic category and life history. Proc R Soc B Biol Sci. 2008;275:1803–9. https://doi.org/10.1098/rspb.2008.0216.
Brothers EB, Williams DMcB, Sale PF. Length of larval life in twelve families of fishes at “One Tree Lagoon”, Great Barrier Reef, Australia. Mar Biol. 1983;76:319–324; https://doi.org/10.1007/BF00393035
Carducci F, Barucca M, Canapa A, Biscotti MA. Rex retroelements and teleost genomes: an overview. Int J Mol Sci. 2018;19:1–15. https://doi.org/10.3390/ijms19113653.
Ezaz T, Berra T, Graves J. Karyotype of the Australian nurseryfish, Kurtus gulliveri (Kurtidae: Perciformes). IAHS Proc Rep. 2006;9:85–8.
Fricke R, Eschmeyer WN, Van der Laan R. Eschmeyer’s catalog of fishes: genera, species, references. Available in http://researcharchive.calacademy.org/research/ichthyology/catalog/fishcatmain.asp (Electronic version accessed 24 July 2022).
Froese R, Pauly D. FishBase, World Wide Web electronic publication, available in https://www.fishbase.org (Electronic version accessed June 15 Dec 2022).
Galetti PM Jr, Aguilar CT, Molina WF. An overview of marine fish cytogenetics. Hydrobiologia. 2000;420:55–62. https://doi.org/10.1023/A:1003977418900.
Gerlach G, Atema J, Raupach MJ, Deister F, Müller A, Kingsford MJ. Cryptic species of cardinalfish with evidence for old and new divergence. Coral Reefs. 2016;35:437–50.
Getlekha N, Molina WF, Cioffi MB, Yano CF, Maneechot N, Bertollo LAC, Supiwong W, Tanomtong A. Repetitive DNAs highlight the role of chromosomal fusions in the karyotype evolution of Dascyllus species (Pomacentridae, Perciformes). Genetica. 2016;144:203–11. https://doi.org/10.1007/s10709-016-9890-5.
Gold JR, Li YC, Shipley NS, Powers PK. Improved methods for working with fish chromosomes with a review of metaphase chromosome banding. J Fish Biol. 1990;37:563–75. https://doi.org/10.1111/j.1095-8649.1990.tb05889.x.
Gornung E. Twenty years of physical mapping of major ribosomal RNA genes across the teleosts: A review of research. Cytogenet Genome Res. 2013;141:90–102. https://doi.org/10.1159/000354832.
Gotoh RO, Sekimoto H, Chiba SN, Hanzawa N. Peripatric differentiation among adjacent marine lake and lagoon populations of a coastal fish, Sphaeramia orbicularis (Apogonidae, Perciformes, Teleostei). Genes Genet Syst. 2009;84:287–95. https://doi.org/10.1266/ggs.84.287.
Gould AL, Henderson JB, Lam AW. Chromosome-level genome assembly of the bioluminescent cardinalfish Siphamia tubifer: An emerging model for symbiosis research. Genome Biol Evol. 2022;14:evac044; https://doi.org/10.1093/gbe/evac044
Groover EM, DiMaggio M, Cassiano EJ. Overview of commonly cultured marine ornamental fish. EDIS. 2020;3:7–7.
Hastie ND, Allshire RC. Human telomeres: fusion and interstitial sites. Trends Genet. 1989;5:326–31. https://doi.org/10.1016/0168-9525(89)90137-6.
Hett AK, Nirchio M, Oliveira C, Siccha ZR, Rossi AR, Sola L. Karyotype characterization of Mugil incilis hancock, 1830 (Mugiliformes: Mugilidae), including a description of an unusual co-localization of major and minor ribosomal genes in the family. Neotrop Ichthyol. 2011;1:107–12. https://doi.org/10.1590/S1679-62252011005000005.
Hoffman EA, Kolm N, Berglund A, Arguello JR, Jones AG. Genetic structure in the coral-reef-associated Banggai cardinalfish. Pterapogon kauderni Mol Ecol. 2005;14:1367–75. https://doi.org/10.1111/j.1365-294X.2005.02538.x.
Howell WM, Black DA. Controlled silver staining of nucleolus organizer region with protective colloidal developer: a 1-step method. Experientia. 1980;36:1014–5. https://doi.org/10.1007/BF01953855.
Kasiroek W, Indananda C, Pinthong K, Supiwong W, Pengseng P, Tanomtong A. NOR polymorphism and chromosome analysis of Banggai Cardinalfish, Pterapogon kauderni (Perciformes, Apogonidae). Cytologia. 2017;82:17–23. https://doi.org/10.1508/cytologia.82.17.
Kasiroek W, Phimphan S, Pinthong K, Suwannapoom C, Aiumsumang S, Liehr T, Supiwong W, Tanomtong A. Comparative cytogenomic analysis of Cardinal fishes (Perciformes, Apogonidae) from Thailand. Nucleus. 2022;65:57–66. https://doi.org/10.1007/s13237-021-00352-5.
Kubat Z, Hobza R, Vyskot B, Kejnovsky E. Microsatelite accumulation on the Y chromosome in Silene latifolia. Genome. 2008;51:350–6.
Levan A, Fredga K, Sandberg AA. Nomenclature for centromeric position on chromosomes. Hereditas. 1964;52:201–20.
Li X, Qi J, Corush JB, Chen J, Zhang J. A chromosome-level genome assembly of the Walking goby (Scartelaos histophorus). Front Mar Sci. 2022;9: 966275. https://doi.org/10.3389/fmars.2022.966275.
Lima-Filho PA, Bertollo LAC, Cioffi MB, et al. Karyotype divergence and spreading of 5S rDNA sequences between genomes of two species: Darter and Emerald gobies (Ctenogobius, Gobiidae). Cytogenet Genome Res. 2014;143:197–203. https://doi.org/10.1159/000360492.
Lima-Filho PA, Cioffi MB, Bertollo LAC, Molina WF. Chromosomal and morphological divergences in Atlantic populations of the frillfin goby Bathygobius soporator (Gobiidae, Perciformes). J Exp Mar Biol Ecol. 2012;434:63–70. https://doi.org/10.1016/j.jembe.2012.08.004.
Luehrmann M, Carleton KL, Cortesi F, Cheney KL, Marshall NJ. Cardinalfishes (Apogonidae) show visual system adaptations typical of nocturnally and diurnally active fish. Mol Ecol. 2019;28:3025–41. https://doi.org/10.1111/mec.15102.
Luiz OJ, Mandin JS, Robertson DR, Rocha LA, Wirtz P, Floeter SR. Ecological traits influencing range expansion across large oceanic dispersal barriers: insights from tropical Atlantic reef fishes. Proc Royal Soc B. 2012;279:1033–40. https://doi.org/10.1098/rspb.2011.1525.
Mabuchi K, Okuda N, Nishida M. Molecular phylogeny and stripe pattern evolution in the cardinal fish genus Apogon. Mol Phylogenet Evol. 2006;38:90–9. https://doi.org/10.1016/j.ympev.2005.05.003.
Mabuchi K, Fraser TH, Song H. Revision of the systematics of the cardinalfishes (Percomorpha: Apogonidae) based on molecular analyses and comparative reevaluation of morphological characters. Zootaxa 2014;3846:151–203; https://https://doi.org/10.11646/zootaxa.3846.2.1
Maggiolini FAM, Sanders AD, Shew CJ, Sulovari A, Mao Y, Puig M, Catacchio CR, Dellino M, Palmisano D, Mercuri L, Bitonto M, Porubský D, et al. Single-cell strand sequencing of a macaque genome reveals multiple nested inversions and breakpoint reuse during primate evolution. Genome Res. 2020;30:1680–93. https://doi.org/10.1101/gr.265322.120.
Marnane MJ, Bellwood DR. Diet and nocturnal foraging in cardinalfishes (Apogonidae) at One Tree Reef, Great Barrier Reef. Australia Mar Ecol Prog Ser. 2002;231:261–8. https://doi.org/10.3354/meps231261.
Molina WF, Alves DE, Araújo WC, Martinez PA, Silva MF, Costa GWWF. Performance of human immunostimulating agents in the improvement of fish cytogenetic preparations. Genet Mol Res. 2010;9:1807–14. https://doi.org/10.4238/vol9-3gmr840.
Molina WF, Galetti PM Jr. Robertsonian rearrangements in the reef fish Chromis (Perciformes, Pomacentridae) involving chromosomes bearing 5S rRNA genes. Genet Mol Biol. 2002;25:373–7. https://doi.org/10.1590/S1415-47572002000400004.
Molina WF, Galetti PM Jr. Karyotypic changes associated to the dispersive potential on Pomacentridae (Pisces, Perciformes). J Exp Mar Biol Ecol. 2004;309:109–19. https://doi.org/10.1016/j.jembe.2004.03.011.
Molina WF. Chromosomal changes and stasis in marine fish groups. In: Pisano E, Ozouf-Costaz C, Foresti F, Kapoor BG, editors. Fish Cytogenetics. Enfield: Science Publishers; 2007. p. 69–110.
Mora C, Sale PF. Are population of coral reef fish open or closed? Trends Ecol Evol. 2002;17:422–8. https://doi.org/10.1016/S0169-5347(02)02584-3.
Motta-Neto CC, Cioffi MB, Bertollo LAC, et al. Molecular cytogenetic analysis of Haemulidae fish (Perciformes): evidence of evolutionary conservation. J Exp Mar Biol Ecol. 2011;401:97–100. https://doi.org/10.1016/j.jembe.2011.07.014.
Motta-Neto CCD, Cioffi MB, Costa GWWF, et al. Overview on karyotype stasis in Atlantic grunts (Eupercaria, Haemulidae) and the evolutionary extensions for other marine fish groups. Front Mar Sci. 2019;6:628. https://doi.org/10.3389/fmars.2019.00628.
Murofushi M. A study of karyotype classification and karyotype evolution in marine teleosts. Report of the Mishima Research Institute of Sciences for Living, Nihon University. 1986;9:95–157.
Murofushi M, Oishi M, Nawa N. Karyological studies in Apogon semilineatus. Report of the Mishima Research Institute of Sciences for Living, Nihon University. 1980;3:47–50.
Ndobe S, Handoko K, Wahyudi D, Yasir M. Irawati, Y., Tanod, W.A., Moore, A.M. Monitoring the endemic ornamental fish Pterapogon kauderni in Bokan Kepulauan, Banggai marine protected area, Indonesia. Depik 2020;9:18–31; https://doi.org/10.13170/depik.9.1.15363
Ndobe S, Moore A, Yasir I, Jompa J. Banggai cardinalfish conservation: priorities, opportunities, and risks. IOP Conf. Series: Earth and Environmental Science 2019;253:012033.
Neira FJ. Larval development of the oral brooding cardinalfish Apogon rueppellii (Teleostei: Apogonidae) in Western Australia. Rec Aust Mus. 1991;15:573–84.
Nelson JS, Grande TC, Wilson MVH. Fishes of the World. 5th ed. New York: John Wiley and Sons Inc; 2016.
Ojima Y, Kojima T. Chromosomal polymorphisms in Apogonidae fishes. Proc Jpn Acad Ser B Phys Biol Sci. 1985;61:79–82. https://doi.org/10.2183/pjab.61.79.
Ozouf-Costaz C, Brandt J, Korting C, Pisano E, Bonillo C, Coutanceau J, Volff J. Genome dynamics and chromosomal localization of the non-LTR retrotransposons Rex1 and Rex3 in Antarctic fish. Antarct Sci. 2004;16:51–7. https://doi.org/10.1017/S0954102004001816.
Pagnozzi JM, Ditchfield AD, Yonenaga-Yassuda Y. Mapping the distribution of the interstitial telomeric (TTAGGG)n sequences in eight species of Brazilian marsupials (Didelphidae) by FISH and the correlation with constitutive heterochromatin. Do ITS represent evidence for fusion events in American marsupials? Cytogenet Genome Res. 2002;98:278–284; https://doi.org/10.1159/000071049
Pendás AM, Moran P, Freije JP, Garcia-Vazquez E. Chromosomal mapping and nucleotide sequence of two tandem repeats of Atlantic salmon 5S rDNA. Cytogenet Cell Genet. 1994;67:31–6. https://doi.org/10.1159/000133792.
Pinkel D, Straume T, Gray J. Cytogenetic analysis using quantitative, high sensitivity, fluorescence hybridization. PNAS. 1986;83:2934–2928. https://doi.org/10.1073/pnas.83.9.2934.
Piñeros VJ, Beltrán-López RG, Baldwin CC, Barraza E, Espinoza E, Martínez JE, Domínguez-Domínguez O. Diversification of the genus Apogon (Lacepède, 1801) (Apogonidae: Perciformes) in the tropical eastern Pacific. Mol Phylogenet Evol. 2019;132:232–42. https://doi.org/10.1016/j.ympev.2018.12.010.
Pusack TJ, Christie MR, Johnson DW, Stallings CD, Hixon MA. Spatial and temporal patterns of larval dispersal in a coral-reef fish metapopulation: evidence of variable reproductive success. Mol Ecol. 2014;23:3396–408. https://doi.org/10.1111/mec.12824.
Qumsiyeh MB. Evolution of number and morphology of mammalian chromosomes. J Hered. 1994;85:455–65. https://doi.org/10.1093/oxfordjournals.jhered.a111501.
Rhyne AL, Tlusty MF, Schofield PJ, Kaufman L, Morris JA, Bruckner AW. Revealing the appetite of the marine aquarium fish trade: The volume and biodiversity of fish imported into the United States. PLoS ONE. 2012;7: e35808. https://doi.org/10.1371/journal.pone.0035808.
Rishi KK. A preliminary report on the karyotypes of eighteen marine fishes. Res Bull Panjab Univ. 1973;24:161–2.
Rivlin KA, Dale G, Rachlin JW. Karyotypic analysis of three species of cardinalfish (Apogonidae) and its implications for the taxonomic status of the genera Apogon and Phaeoptyx. Ann N Y Acad Sci. 1986;463:211–3. https://doi.org/10.1111/j.1749-6632.1986.tb21549.x.
Rivlin KA, Rachlin JW, Dale G. Intraspecific chromosomal variation in Apogon binotatus (Perciformes: Apogonidae) from the Florida Keys and St. Croix. Annals of New York Academy of Sciences 1987;494:263–265.
Rivlin KA, Rachlin JW, Warkentine BE. G banding of the chromosomes of Apogon maculatus and A. pseudomaculatus (Perciformes: Apogonidae). Ann N Y Acad Sci. 1988;529:160–163; https://doi.org/10.1111/j.1749-6632.1988.tb51448.x
Rueger T, Harrison HB, Jones GP, Mansour H, Berumen ML. Resolving genealogical relationships in the Pyjama cardinalfish, Sphaeramia nematoptera (Apogonidae) with 23 novel microsatellite markers. Conservation Genet Resour. 2015;7:623–6. https://doi.org/10.1007/s12686-015-0461-3.
Sale PF. The ecology of fishes on coral reefs. California: Academic Press; 1993.
Sena DCS, Molina WF. Chromosomal rearrangements associated with pelagic larval duration in Labridae (Perciformes). J Exp Mar Biol Ecol. 2007;353:203–10. https://doi.org/10.1016/j.jembe.2007.08.020.
Da Silva C, Hadji H, Ozouf-Costaz C, Nicaud S, Jaillon O, Weissenbach J, Crollius HR. Remarkable compartmentalization of transposable elements and pseudogenes in the heterochromatin of the Tetraodon nigroviridis genome. PNAS. 2002;99:1636–41. https://doi.org/10.1073/pnas.202284199.
Silva SAS, Lima-Filho PA, Motta-Neto CC, et al. High chromosomal evolutionary dynamics in sleeper gobies (Eleotridae) and notes on disruptive biological factors in Gobiiformes karyotypes (Osteichthyes, Teleostei). Mar Life Sci Technol. 2021;3:293–302. https://doi.org/10.1007/s42995-020-00084-6.
Sumner AT. A simple technique for demonstrating centromeric heterochromatin. Exp Cell Res. 1972;75:304–6. https://doi.org/10.1016/0014-4827(72)90558-7.
Thacker CE. Patterns of divergence in fish species separated by the Isthmus of Panama. BMC Evol Biol. 2017;17:111. https://doi.org/10.1186/s12862-017-0957-4.
Thacker CE, Roje DM. Phylogeny of cardinalfishes (Teleostei: Gobiiformes: Apogonidae) and the evolution of visceral bioluminescence. Mol Phylogenet Evol. 2009;52:735–45. https://doi.org/10.1016/j.ympev.2009.05.017.
Thacker CE. Species and shape diversification are inversely correlated among gobies and cardinalfishes (Teleostei: Gobiiformes). Org Divers Evol. 2014;14 419–436; https://doi.org/10.1007/s13127-014-0175-5
Tine M, Kuhl H, Gagnaire PA, et al. European sea bass genome and its variation provide insights into adaptation to euryhalinity and speciation. Nat commun. 2014;5:5770. https://doi.org/10.1038/ncomms6770.
Underwood JN, Travers MJ, Snow M, Puotinen M, Gows G. Cryptic lineages in the Wolf Cardinalfish living in sympatry on remote coral atolls. Mol Phylogenet Evol. 2019;132:183–93. https://doi.org/10.1016/j.ympev.2018.12.001.
Valente GT, Mazzuchelli J, Ferreira IA, Poletto AB, Fantinatti BEA, Martins C. Cytogenetic mapping of the retroelements Rex1, Rex3 and Rex6 among cichlid fish: new insights on the chromosomal distribution of transposable elements. Cytogenet Genome Res. 2011;133:34–42. https://doi.org/10.1159/000322888.
Vasil’ev VP, Grigoryan KA. Chromosome polymorphism and karyological relationships in the group of gobies Neogobius cephalarges Pallas – Neogobius platyrostris Pallas (Gobiidae). Genetics 1994;9:1251–1259.
White TJ, Bruns T, Lee S, Taylor J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Shinsky JJ, White TJ, editors. PCR protocols: a guide to methods and applications. San Diego: Academic Press Inc; 1990. p. 315–22.
Acknowledgements
The authors would like to thank the National Council for Scientific and Technological Development (CNPq) for financial assistance (Grant number 442664/2015-0; 442626/2019-3 and 301458/2019-7), and to the Instituto Brasileiro do Meio Ambiente e dos Recursos Naturais Renováveis (IBAMA) for the license to collect the specimens (Processes number 19135-8, #131360-1 and #27027-2) and to Federal University of Rio Grande do Norte (UFRN), for supporting this study. We also thank José Garcia Júnior for species identification.
Author information
Authors and Affiliations
Contributions
Conceptualization, WFM; Methodology, EWS, GWWFC, VCSO; Formal analysis, EWS; Investigation, EWS, VCSO, GWWFC and WFM; Writing—Original Draft, EWS, and WFM; Writing—Review and Editing, MBC, LACB, GWWFC, KDJA, and WFM; Funding acquisition and Project administration, WFM.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Institutional review board statement
The study was conducted according to the guidelines of the Declaration of Helsinki and approved by the Ethics Committee on the use of animals in research of Universidade Federal do Rio Grande do Norte (License # 44/2015).
Additional information
Corresponding Editors and Reviewers: Rabindra Nath Chatterjee, Umesh C. Lavania
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
dos Santos, É.W., de Bello Cioffi, M., da Costa, G.W.W.F. et al. The diploid number decrease in cardinalfishes (Apogonidae, Kurtiformes): chromosomal rearrangements and related biological features. Nucleus (2023). https://doi.org/10.1007/s13237-023-00438-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13237-023-00438-2