Abstract
Purpose
To predict prognosis in HIV-negative cryptococcal meningitis (CM) patients by developing and validating a machine learning (ML) model.
Methods
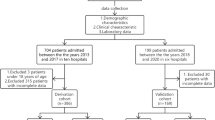
This study involved 523 HIV-negative CM patients diagnosed between January 1, 1998, and August 31, 2022, by neurologists from 3 tertiary Chinese centers. Prognosis was evaluated at 10 weeks after the initiation of antifungal therapy.
Results
The final prediction model for HIV-negative CM patients comprised 8 variables: Cerebrospinal fluid (CSF) cryptococcal count, CSF white blood cell (WBC), altered mental status, hearing impairment, CSF chloride levels, CSF opening pressure (OP), aspartate aminotransferase levels at admission, and decreased rate of CSF cryptococcal count within 2 weeks after admission. The areas under the curve (AUCs) in the internal, temporal, and external validation sets were 0.87 (95% CI 0.794–0.944), 0.92 (95% CI 0.795–1.000), and 0.86 (95% CI 0.744–0.975), respectively. An artificial intelligence (AI) model was trained to detect and count cryptococci, and the mean average precision (mAP) was 0.993.
Conclusion
A ML model for predicting prognosis in HIV-negative CM patients was built and validated, and the model might provide a reference for personalized treatment of HIV-negative CM patients. The change in the CSF cryptococcal count in the early phase of HIV-negative CM treatment can reflect the prognosis of the disease. In addition, utilizing AI to detect and count CSF cryptococci in HIV-negative CM patients can eliminate the interference of human factors in detecting cryptococci in CSF samples and reduce the workload of the examiner.




Similar content being viewed by others
Data availability
The datasets used and analyzed in this study are available from the corresponding author on reasonable request.
Abbreviations
- CM :
-
Cryptococcal meningitis
- CSF :
-
Cerebrospinal fluid
- WBC :
-
White blood cell
- ML :
-
Machine learning
- AI :
-
Artificial intelligence
- TRIPOD :
-
Transparent Reporting of a Multivariable Prediction Model for Individual Prognosis or Diagnosis
- OP :
-
Opening pressure
- BMI :
-
Body mass index
- AUC :
-
Area under the curve
- MAP :
-
Mean average precision
- IOU :
-
Intersection over union
- PR :
-
Precision recall
- LR :
-
Logistic regression
- WSI :
-
Whole-slide images
- IQR :
-
Interquartile range
- LASSO :
-
Least absolute shrinkage and the selection operator
- RF :
-
Random forest
- KNN :
-
k-Nearest neighbours
- DT :
-
Decision tree
- EFA :
-
Early fungicidal activity
- CFU :
-
Colony-forming-units
References
Francisco EC, de Jong AW, Hagen F (2021) Cryptococcosis and cryptococcus. Mycopathologia 186(5):729–731
Williamson PR, Jarvis JN, Panackal AA, Fisher MC, Molloy SF, Loyse A, Harrison TS (2017) Cryptococcal meningitis: epidemiology, immunology, diagnosis and therapy. Nat Rev Neurol 13(1):13–24
Brizendine KD, Baddley JW, Pappas PG (2013) Predictors of mortality and differences in clinical features among patients with cryptococcosis according to immune status. PLoS ONE 8(3):e60431
Bratton EW, El Husseini N, Chastain CA, Lee MS, Poole C, Sturmer T, Juliano JJ, Weber DJ, Perfect JR (2012) Comparison and temporal trends of three groups with cryptococcosis: HIV-infected, solid organ transplant, and HIV-negative/non-transplant. PLoS ONE 7(8):e43582
Rathore SS, Sathiyamoorthy J, Lalitha C, Ramakrishnan J (2022) A holistic review on Cryptococcus neoformans. Microb Pathog 166:105521
Motoa G, Pate A, Chastain D, Mann S, Canfield GS, Franco-Paredes C, Henao-Martinez AF (2020) Increased cryptococcal meningitis mortality among HIV negative, non-transplant patients: a single US center cohort study. Ther Adv Infect Dis 7:2049936120940881
Subudhi S, Verma A, Patel AB, Hardin CC, Khandekar MJ, Lee H, McEvoy D, Stylianopoulos T, Munn LL, Dutta S et al (2021) Comparing machine learning algorithms for predicting ICU admission and mortality in COVID-19. NPJ Digit Med 4(1):87
Yang Y, Xu L, Sun L, Zhang P, Farid SS (2022) Machine learning application in personalised lung cancer recurrence and survivability prediction. Comput Struct Biotechnol J 20:1811–1820
Mfateneza E, Rutayisire PC, Biracyaza E, Musafiri S, Mpabuka WG (2022) Application of machine learning methods for predicting infant mortality in Rwanda: analysis of Rwanda demographic health survey 2014–15 dataset. BMC Pregnancy Childbirth 22(1):388
Litjens G, Kooi T, Bejnordi BE, Setio AAA, Ciompi F, Ghafoorian M, van der Laak J, van Ginneken B, Sanchez CI (2017) A survey on deep learning in medical image analysis. Med Image Anal 42:60–88
Marzi C, d’Ambrosio A, Diciotti S, Bisecco A, Altieri M, Filippi M, Rocca MA, Storelli L, Pantano P, Tommasin S, Cortese R, De Stefano N, Tedeschi G, Gallo A; INNI Network (2022) Prediction of the information processing speed performance in multiple sclerosis using a machine learning approach in a large multicenter magnetic resonance imaging data set. Hum Brain Mapp 44(1):186–202
Hwangbo L, Kang YJ, Kwon H, Lee JI, Cho HJ, Ko JK, Sung SM, Lee TH (2022) Stacking ensemble learning model to predict 6-month mortality in ischemic stroke patients. Sci Rep 12(1):17389
Hakeem H, Feng W, Chen Z, Choong J, Brodie MJ, Fong SL, Lim KS, Wu J, Wang X, Lawn N et al (2022) Development and validation of a deep learning model for predicting treatment response in patients with newly diagnosed epilepsy. JAMA Neurol 79(10):986–996
Vidal JE, Gerhardt J, Peixoto de Miranda EJ, Dauar RF, Oliveira Filho GS, Penalva de Oliveira AC, Boulware DR (2012) Role of quantitative CSF microscopy to predict culture status and outcome in HIV-associated cryptococcal meningitis in a Brazilian cohort. Diagn Microbiol Infect Dis 73(1):68–73
Saag MS, Powderly WG, Cloud GA, Robinson P, Grieco MH, Sharkey PK, Thompson SE, Sugar AM, Tuazon CU, Fisher JF et al (1992) Comparison of amphotericin B with fluconazole in the treatment of acute AIDS-associated cryptococcal meningitis. The NIAID Mycoses Study Group and the AIDS Clinical Trials Group. N Engl J Med 326(2):83–89
Zhang C, Tan Z, Tian F (2020) Impaired consciousness and decreased glucose concentration of CSF as prognostic factors in immunocompetent patients with cryptococcal meningitis. BMC Infect Dis 20(1):69
Bicanic T, Muzoora C, Brouwer AE, Meintjes G, Longley N, Taseera K, Rebe K, Loyse A, Jarvis J, Bekker LG et al (2009) Independent association between rate of clearance of infection and clinical outcome of HIV-associated cryptococcal meningitis: analysis of a combined cohort of 262 patients. Clin Infect Dis 49(5):702–709
Liang H, Tsui BY, Ni H, Valentim CCS, Baxter SL, Liu G, Cai W, Kermany DS, Sun X, Chen J et al (2019) Evaluation and accurate diagnoses of pediatric diseases using artificial intelligence. Nat Med 25(3):433–438
Collins GS, Reitsma JB, Altman DG, Moons KG (2015) Transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD). Ann Intern Med 162(10):735–736
Zhang K, Li H, Zhang L, Liao W, Ling L, Li X, Lin J, Xu B, Pan W, Zhang Q (2020) Cerebrospinal fluid microscopy as an index for predicting the prognosis of cryptococcal meningitis patients with and without HIV. Future Microbiol 15:1645–1652
Perfect JR, Dismukes WE, Dromer F, Goldman DL, Graybill JR, Hamill RJ, Harrison TS, Larsen RA, Lortholary O, Nguyen MH et al (2010) Clinical practice guidelines for the management of cryptococcal disease: 2010 update by the infectious diseases society of america. Clin Infect Dis 50(3):291–322
Xu L, Liu J, Zhang Q, Li M, Liao J, Kuang W, Zhu C, Yi H, Peng F (2018) Triple therapy versus amphotericin B plus flucytosine for the treatment of non-HIV- and non-transplant-associated cryptococcal meningitis: retrospective cohort study. Neurol Res 40(5):398–404
Liu ZY, Wang GQ, Zhu LP, Lyu XJ, Zhang QQ, Yu YS, Zhou ZH, Liu YB, Cai WP, Li RY et al (2018) Expert consensus on the diagnosis and treatment of cryptococcal meningitis. Zhonghua Nei Ke Za Zhi 57(5):317–323
Segal BH, Herbrecht R, Stevens DA, Ostrosky-Zeichner L, Sobel J, Viscoli C, Walsh TJ, Maertens J, Patterson TF, Perfect JR et al (2008) Defining responses to therapy and study outcomes in clinical trials of invasive fungal diseases: Mycoses Study Group and European Organization for Research and Treatment of Cancer consensus criteria. Clin Infect Dis 47(5):674–683
Li Z, Liu Y, Chong Y, Li X, Jie Y, Zheng X, Yan Y (2019) Fluconazole plus flucytosine is a good alternative therapy for non-HIV and non-transplant-associated cryptococcal meningitis: a retrospective cohort study. Mycoses 62(8):686–691
Simon N, Friedman J, Hastie T, Tibshirani R (2011) Regularization paths for Cox’s proportional hazards model via coordinate descent. J Stat Softw 39(5):1–13
R Core Team (2019) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. https://www.R-project.org/
Jarvis JN, Bicanic T, Loyse A, Namarika D, Jackson A, Nussbaum JC, Longley N, Muzoora C, Phulusa J, Taseera K et al (2014) Determinants of mortality in a combined cohort of 501 patients with HIV-associated cryptococcal meningitis: implications for improving outcomes. Clin Infect Dis 58(5):736–745
Mora DJ, da Cunha Colombo ER, Ferreira-Paim K, Andrade-Silva LE, Nascentes GA, Silva-Vergara ML (2012) Clinical, epidemiological and outcome features of patients with cryptococcosis in Uberaba, Minas Gerais, Brazil. Mycopathologia 173(5–6):321–327
Pitisuttithum P, Tansuphasawadikul S, Simpson AJ, Howe PA, White NJ (2001) A prospective study of AIDS-associated cryptococcal meningitis in Thailand treated with high-dose amphotericin B. J Infect 43(4):226–233
Brouwer AE, Rajanuwong A, Chierakul W, Griffin GE, Larsen RA, White NJ, Harrison TS (2004) Combination antifungal therapies for HIV-associated cryptococcal meningitis: a randomised trial. Lancet 363(9423):1764–1767
Dromer F, Mathoulin-Pelissier S, Launay O, Lortholary O, French Cryptococcosis Study G (2007) Determinants of disease presentation and outcome during cryptococcosis: the CryptoA/D study. PLoS Med 4(2):e21
de Oliveira L, Melhem MSC, Buccheri R, Chagas OJ, Vidal JE, Diaz-Quijano FA (2022) Early clinical and microbiological predictors of outcome in hospitalized patients with cryptococcal meningitis. BMC Infect Dis 22(1):138
Julin Gu HW, Liao W (2002) Study on the viability of cryptococcus in cerebrospinal fluid of patients with cryptococcal meningitis. Chin J Dermatol 35(5):372–373
Diamond RD, Bennett JE (1974) Prognostic factors in cryptococcal meningitis. A study in 111 cases. Ann Intern Med 80(2):176–181
Mitchell TG, Perfect JR (1995) Cryptococcosis in the era of AIDS–100 years after the discovery of Cryptococcus neoformans. Clin Microbiol Rev 8(4):515–548
Abassi M, Boulware DR, Rhein J (2015) Cryptococcal meningitis: diagnosis and management update. Curr Trop Med Rep 2(2):90–99
Graybill JR, Sobel J, Saag M, van Der Horst C, Powderly W, Cloud G, Riser L, Hamill R, Dismukes W (2000) Diagnosis and management of increased intracranial pressure in patients with AIDS and cryptococcal meningitis. The NIAID Mycoses Study Group and AIDS Cooperative Treatment Groups. Clin Infect Dis 30(1):47–54
van der Horst CM, Saag MS, Cloud GA, Hamill RJ, Graybill JR, Sobel JD, Johnson PC, Tuazon CU, Kerkering T, Moskovitz BL et al (1997) Treatment of cryptococcal meningitis associated with the acquired immunodeficiency syndrome National Institute of Allergy and Infectious Diseases Mycoses Study Group and AIDS Clinical Trials Group. N Engl J Med 337(1):15–21
Acknowledgements
The authors thank Jianmin Li and Weiwei Gao of Unicom (Guangdong) Industrial internet Co., Ltd. for their contribution to the webpage construction of the prediction model and AI-assisted cryptococcal counting in this study.
Funding
The study was supported by the National Science Foundation of China (No. 82071265).
Author information
Authors and Affiliations
Contributions
Junyu Liu, Yaxin Lu, Ying Jiang, Zifeng Liu, and Fuhua Peng contributed to the study design. Fuhua Peng, Ying Jiang, Junyu Liu, Jia Liu, and Jiayin Liang did the literature search. Yaxin Lu and Zifeng Liu analyzed the data and created figures and tables. Junyu Liu wrote the first draft. Junyu Liu, Qilong Zhang, Hua Li, Xiufeng Zhong, Hui Bu, Zhanhang Wang, Liuxu Fan, Panpan Liang, Jia Xie, Yuan Wang, Jiayin Gong, Haiying Chen, Yangyang Dai, Lu Yang, Xiaohong Su, Anni Wang, Lei Xiong, and Han Xia contributed to the data collection. All authors contributed to the interpretation of results, reviewed and critically revised the manuscript, and approved the final version for submission.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
All procedures performed in studies involving the patient included were in accordance with the ethical standards of the institutional board and with the 1964 Helsinki Declaration. This study is approved by the Medical Ethics Committee of the Third Affiliated Hospital of Sun Yat-sen University (approval no. [2022] 02-166-01). At admission, the subjects or the guardians of patients with cognitive impairment provided written informed consent for research and publication.
Consent for publication
We have obtained consent to publish from the participant to report individual patient data.
Conflict of interest
The authors declare that they have no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.

Supplementary Fig. 1
Correlation coefficient chart between variables. (PNG 555 kb)

Supplementary Fig. 2
Feature selection using least absolute shrinkage and the selection operator (LASSO) regression model. (PNG 178 kb)

Supplementary Fig. 3
Calibration curve: (A)internal validation set, (B)temporal validation set, (C)external validation set. (PNG 339 kb)

Supplementary Fig. 4
Online webpage: (A) cryptococcal automatic count, (B) Calculator for risk prediction model. (PNG 1528 kb)
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Liu, J., Lu, Y., Liu, J. et al. Development and validation of a machine learning model to predict prognosis in HIV-negative cryptococcal meningitis patients: a multicenter study. Eur J Clin Microbiol Infect Dis 42, 1183–1194 (2023). https://doi.org/10.1007/s10096-023-04653-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10096-023-04653-2