Abstract
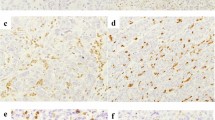
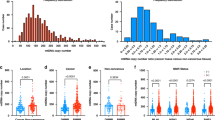
Deficient mismatch repair (dMMR)/microsatellite instability (MSI) colorectal cancer (CRC) has high immunogenicity and better prognosis compared with proficient MMR (pMMR)/microsatellite stable (MSS) CRC. Although the activation of the cyclic GMP-AMP synthase (cGAS)-stimulator of interferon genes (STING) pathway has been considered to contribute to the high number of CD8+ TILs, its role in dMMR/MSI CRC is largely unknown. In this study, to examine the role of the cGAS-STING pathway on the recruitment of CD8+ TILs in dMMR/MSI CRC, we used public datasets and clinical tissue samples in our cohorts to evaluate the expression of cGAS, STING, and CD8+ TILs in pMMR/MSS and dMMR/MSI CRCs. According to the analysis of public datasets, the expression of cGAS-STING, CD8 effector gene signature, and CXCL10-CCL5, chemoattractants for CD8+ TILs which regulated by the cGAS-STING pathway, was significantly upregulated in dMMR/MSI CRC, and the expression of cGAS-STING was significantly associated with the expression of CD8 effector gene signature. Immunohistochemistry staining of the clinical tissue samples (n = 283) revealed that cGAS-STING was highly expressed in tumor cells of dMMR CRC, and higher expression of cGAS-STING in tumor cells was significantly associated with the increased number of CD8+ TILs. Moreover, we demonstrated that the downregulation of MMR gene in human CRC cell lines enhanced the activation of the cGAS-STING pathway. Taken together, for the first time, we found that dMMR/MSI CRC has maintained a high level of cGAS-STING expression in tumor cells, which might contribute to abundant CD8+ TILs and immune-active TME.



Similar content being viewed by others
Data availability
The datasets generated during and/or analyzed during the current study are available from corresponding author on reasonable request.
Abbreviations
- CCL5:
-
C–C chemokine ligand-5
- cGAMP:
-
Cyclic GMP-AMP
- cGAS:
-
Cyclic GMP-AMP synthase,
- CIN:
-
Chromosomal instability
- CRC:
-
Colorectal cancer
- CXCL10:
-
C-X-C motif ligand 10
- dMMR:
-
Mismatch repair deficient
- ICI:
-
Immune checkpoint inhibitor
- IRF3:
-
Interferon regulatory factor 3
- MLH1:
-
MutL homolog 1
- MSH2:
-
MutS homolog 2
- MSH6:
-
MutS homolog 6
- MSI:
-
Microsatellite instability
- MSS:
-
Microsatellite stable
- pMMR:
-
Mismatch repair proficient
- PMS2:
-
PMS1 homolog 2
- STING:
-
Stimulator of interferon genes
- TREX1:
-
Three-prime repair exonuclease 1
References
Siegel RL, Miller KD, Goding Sauer A, Fedewa SA, Butterly LF, Anderson JC, Cercek A, Smith RA, Jemal A (2020) Colorectal cancer statistics, 2020. CA Cancer J Clin 70:145–164. https://doi.org/10.3322/caac.21601
Keum N, Giovannucci E (2019) Global burden of colorectal cancer: emerging trends, risk factors and prevention strategies. Nat Rev Gastroenterol Hepatol 16:713–732. https://doi.org/10.1038/s41575-019-0189-8
Xie YH, Chen YX, Fang JY (2020) Comprehensive review of targeted therapy for colorectal cancer. Signal Transduct Target Ther 5:22. https://doi.org/10.1038/s41392-020-0116-z
Tutlewska K, Lubinski J, Kurzawski G (2013) Germline deletions in the EPCAM gene as a cause of Lynch syndrome - literature review. Hered Cancer Clin Pract 11:9. https://doi.org/10.1186/1897-4287-11-9
Kuiper RP, Vissers LE, Venkatachalam R et al (2011) Recurrence and variability of germline EPCAM deletions in Lynch syndrome. Hum Mutat 32:407–414. https://doi.org/10.1002/humu.21446
Amodio V, Mauri G, Reilly NM, Sartore-Bianchi A, Siena S, Bardelli A, Germano G (2021) Mechanisms of immune escape and resistance to checkpoint inhibitor therapies in mismatch repair deficient metastatic colorectal cancers. Cancers (Basel). https://doi.org/10.3390/cancers13112638
Randrian V, Evrard C, Tougeron D (2021) Microsatellite instability in colorectal cancers: carcinogenesis, neo-antigens immuno-resistance and emerging therapies. Cancers (Basel). https://doi.org/10.3390/cancers13123063
Boland CR, Goel A (2010) Microsatellite instability in colorectal cancer. Gastroenterology 138:2073-2087.e2073. https://doi.org/10.1053/j.gastro.2009.12.064
Deshpande M, Romanski PA, Rosenwaks Z, Gerhardt J (2020) Gynecological cancers caused by deficient mismatch repair and microsatellite instability. Cancers (Basel) 12:3319. https://doi.org/10.3390/cancers12113319
Pritchard CC, Grady WM (2011) Colorectal cancer molecular biology moves into clinical practice. Gut 60:116–129. https://doi.org/10.1136/gut.2009.206250
Phillips SM, Banerjea A, Feakins R, Li SR, Bustin SA, Dorudi S (2004) Tumour-infiltrating lymphocytes in colorectal cancer with microsatellite instability are activated and cytotoxic. Br J Surg 91:469–475. https://doi.org/10.1002/bjs.4472
Ganesh K, Stadler ZK, Cercek A, Mendelsohn RB, Shia J, Segal NH, Diaz LA Jr (2019) Immunotherapy in colorectal cancer: rationale, challenges and potential. Nat Rev Gastroenterol Hepatol 16:361–375. https://doi.org/10.1038/s41575-019-0126-x
Le DT, Durham JN, Smith KN et al (2017) Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science 357:409–413. https://doi.org/10.1126/science.aan6733
Wu J, Sun L, Chen X, Du F, Shi H, Chen C, Chen ZJ (2013) Cyclic GMP-AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science 339:826–830. https://doi.org/10.1126/science.1229963
Gao M, He Y, Tang H, Chen X, Liu S, Tao Y (2020) cGAS/STING: novel perspectives of the classic pathway. Mol Biomed. https://doi.org/10.1186/s43556-020-00006-z
Li T, Chen ZJ (2018) The cGAS-cGAMP-STING pathway connects DNA damage to inflammation, senescence, and cancer. J Exp Med 215:1287–1299. https://doi.org/10.1084/jem.20180139
Jiang M, Chen P, Wang L et al (2020) cGAS-STING, an important pathway in cancer immunotherapy. J Hematol Oncol 13:81. https://doi.org/10.1186/s13045-020-00916-z
Tokunaga R, Zhang W, Naseem M, Puccini A, Berger MD, Soni S, McSkane M, Baba H, Lenz H-J (2018) CXCL9, CXCL10, CXCL11/CXCR3 axis for immune activation–a target for novel cancer therapy. Cancer Treat Rev 63:40–47. https://doi.org/10.1016/j.ctrv.2017.11.007
Sokolowska O, Nowis D (2018) STING signaling in cancer cells: Important or not? Arch Immunol Ther Exp (Warsz) 66:125–132. https://doi.org/10.1007/s00005-017-0481-7
Song S, Peng P, Tang Z et al (2017) Decreased expression of STING predicts poor prognosis in patients with gastric cancer. Sci Rep 7:39858. https://doi.org/10.1038/srep39858
Chon HJ, Kim H, Noh JH et al (2019) STING signaling is a potential immunotherapeutic target in colorectal cancer. J Cancer 10:4932–4938. https://doi.org/10.7150/jca.32806
Bu Y, Liu F, Jia Q-A, Yu S-N (2016) Decreased expression of TMEM173 predicts poor prognosis in patients with hepatocellular carcinoma. PLoS One 11:e0165681. https://doi.org/10.1371/journal.pone.0165681
Xia T, Konno H, Ahn J, Barber GN (2016) Deregulation of STING signaling in colorectal carcinoma constrains DNA damage responses and correlates with tumorigenesis. Cell Rep 14:282–297. https://doi.org/10.1016/j.celrep.2015.12.029
Bakhoum SF, Cantley LC (2018) The multifaceted role of chromosomal instability in cancer and its microenvironment. Cell 174:1347–1360. https://doi.org/10.1016/j.cell.2018.08.027
de la Chapelle A, Hampel H (2010) Clinical relevance of microsatellite instability in colorectal cancer. J Clin Oncol 28:3380–3387. https://doi.org/10.1200/jco.2009.27.0652
Dou Z, Ghosh K, Vizioli MG et al (2017) Cytoplasmic chromatin triggers inflammation in senescence and cancer. Nature 550:402–406. https://doi.org/10.1038/nature24050
Santaguida S, Richardson A, Iyer DR et al (2017) Chromosome mis-segregation generates cell-cycle-arrested cells with complex karyotypes that are eliminated by the immune system. Dev Cell 41:638-651.e635. https://doi.org/10.1016/j.devcel.2017.05.022
Lu C, Guan J, Lu S et al (2021) DNA sensing in mismatch repair-deficient tumor cells is essential for anti-tumor immunity. Cancer Cell 39(96–108):e106. https://doi.org/10.1016/j.ccell.2020.11.006
Guan J, Lu C, Jin Q et al (2021) MLH1 deficiency-triggered DNA hyperexcision by exonuclease 1 activates the cGAS-STING pathway. Cancer Cell 39(109–121):e105. https://doi.org/10.1016/j.ccell.2020.11.004
Mowat C, Mosley SR, Namdar A, Schiller D, Baker K (2021) Anti-tumor immunity in mismatch repair-deficient colorectal cancers requires type I IFN–driven CCL5 and CXCL10. J Exp Med. https://doi.org/10.1084/jem.20210108
Gao J, Aksoy BA, Dogrusoz U et al (2013) Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. https://doi.org/10.1126/scisignal.2004088
Davoli T, Uno H, Wooten EC, Elledge SJ (2017) Tumor aneuploidy correlates with markers of immune evasion and with reduced response to immunotherapy. Science. https://doi.org/10.1126/science.aaf8399
Zhang C, Li J, Wang H, Song SW (2016) Identification of a five B cell-associated gene prognostic and predictive signature for advanced glioma patients harboring immunosuppressive subtype preference. Oncotarget 7:73971–73983. https://doi.org/10.18632/oncotarget.12605
Herbst RS, Baas P, Kim DW et al (2016) Pembrolizumab versus docetaxel for previously treated, PD-L1-positive, advanced non-small-cell lung cancer (KEYNOTE-010): a randomised controlled trial. Lancet 387:1540–1550. https://doi.org/10.1016/s0140-6736(15)01281-7
Kikuchi T, Mimura K, Okayama H et al (2019) A subset of patients with MSS/MSI-low-colorectal cancer showed increased CD8(+) TILs together with up-regulated IFN-γ. Oncol Lett 18:5977–5985. https://doi.org/10.3892/ol.2019.10953
Noda M, Okayama H, Tachibana K et al (2018) glycosyltransferase gene expression identifies a poor prognostic colorectal cancer subtype associated with mismatch repair deficiency and incomplete glycan synthesis. Clin Cancer Res 24:4468–4481. https://doi.org/10.1158/1078-0432.ccr-17-3533
Abe T, Harashima A, Xia T, Konno H, Konno K, Morales A, Ahn J, Gutman D, Barber GN (2013) STING recognition of cytoplasmic DNA instigates cellular defense. Mol Cell 50:5–15. https://doi.org/10.1016/j.molcel.2013.01.039
Zumwalt TJ, Arnold M, Goel A, Boland CR (2015) Active secretion of CXCL10 and CCL5 from colorectal cancer microenvironments associates with GranzymeB+ CD8+ T-cell infiltration. Oncotarget 6:2981–2991. https://doi.org/10.18632/oncotarget.3205
Aldinucci D, Colombatti A (2014) The inflammatory chemokine CCL5 and cancer progression. Med Inflamm 2014:292376. https://doi.org/10.1155/2014/292376
Yang L, Wang B, Qin J, Zhou H, Majumdar AP, Peng F (2018) Blockade of CCR5-mediated myeloid derived suppressor cell accumulation enhances anti-PD1 efficacy in gastric cancer. Immunopharmacol Immunotoxicol 40:91–97
Hemphill WO, Simpson SR, Liu M, Salsbury FR Jr, Hollis T, Grayson JM, Perrino FW (2021) TREX1 as a novel immunotherapeutic target. Front Immunol 12:660184. https://doi.org/10.3389/fimmu.2021.660184
Sledz CA, Holko M, de Veer MJ, Silverman RH, Williams BR (2003) Activation of the interferon system by short-interfering RNAs. Nat Cell Biol 5:834–839. https://doi.org/10.1038/ncb1038
Konno H, Yamauchi S, Berglund A, Putney RM, Mulé JJ, Barber GN (2018) Suppression of STING signaling through epigenetic silencing and missense mutation impedes DNA damage mediated cytokine production. Oncogene 37:2037–2051. https://doi.org/10.1038/s41388-017-0120-0
Schoggins JW, Wilson SJ, Panis M, Murphy MY, Jones CT, Bieniasz P, Rice CM (2011) A diverse range of gene products are effectors of the type I interferon antiviral response. Nature 472:481–485. https://doi.org/10.1038/nature09907
Flood BA, Higgs EF, Li S, Luke JJ, Gajewski TF (2019) STING pathway agonism as a cancer therapeutic. Immunol Rev 290:24–38. https://doi.org/10.1111/imr.12765
Wilson R, Espinosa-Diez C, Kanner N et al (2016) MicroRNA regulation of endothelial TREX1 reprograms the tumour microenvironment. Nat Commun 7:13597. https://doi.org/10.1038/ncomms13597
Demaria O, De Gassart A, Coso S et al (2015) STING activation of tumor endothelial cells initiates spontaneous and therapeutic antitumor immunity. Proc Natl Acad Sci U S A 112:15408–15413. https://doi.org/10.1073/pnas.1512832112
Zhu Y, An X, Zhang X, Qiao Y, Zheng T, Li X (2019) STING: a master regulator in the cancer-immunity cycle. Mol Cancer 18:152. https://doi.org/10.1186/s12943-019-1087-y
Acknowledgements
This work was supported by the grants from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (to S. Nakajima and K. Kono). We thank Masayo Sugeno, Sakino Arai, Eri Takahashi, and Saori Naruse for excellent technical assistant and helpful secretarial assistance.
Author information
Authors and Affiliations
Contributions
SN and KK contributed to the study conception. SN, HO, KM, and KK contributed to the supervision of the study. Material preparation was performed by AK, SN, and KS. Data curation and analysis were performed by SN, AK, and HO. Data collection and investigation were performed by AK, SN, TMa, KS, TK, EE. Patient recruitment was carried out by AK, HO, TMa, TK, EE, MI, YK, MS, ZS, SF, WS, HO, TMo, and SO. The first draft and the revised version of the manuscript were written by AK, SN, and KK. All authors read and approved the manuscript.
Corresponding author
Ethics declarations
Conflict of Interest
No potential conflicts of interest relevant to this article were reported.
Ethics approval
This study was conducted in accordance with the principles of the Declaration of Helsinki and was approved by the Institutional Review Board of Fukushima Medical University (Fukushima, Japan). Cell line authentication: Human colorectal cancer cell lines were purchased from the Korean Cell Line Bank (SNU81) and the American Type Culture Collection (SW480), and the cell lines have been authenticated using STR analysis (Promega Japan, Tokyo, Japan).
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Kaneta, A., Nakajima, S., Okayama, H. et al. Role of the cGAS-STING pathway in regulating the tumor-immune microenvironment in dMMR/MSI colorectal cancer. Cancer Immunol Immunother 71, 2765–2776 (2022). https://doi.org/10.1007/s00262-022-03200-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00262-022-03200-w