Abstract
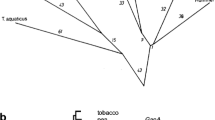
Angiosperms and algae possess two distinct glyceraldehyde-3-phosphate dehydrogenase (GAPDH) enzymes, an NAD+-dependent tetramer involved in cytosolic glycolysis and an NADP+-dependent enzyme of the Calvin cycle in chloroplasts. We have found that the gymnosperm Pinus sylvestris possesses, in addition to these, a nuclear-encoded, plastid-specific, NAD+-dependent GAPDH, designated GapCp, which has not previously been described from any plant. Several independent full-size cDNAs for this enzyme were isolated which encode a functional transit peptide and mature subunit very similar to that of cytosolic GAPDH of angiosperms and algae. A molecular phylogeny reveals that chloroplast GapCp and cytosolic GapC arose through gene duplication early in chlorophyte evolution. The GapCp gene is expressed as highly as that for GapC in light-grown pine seedlings. These findings suggest that aspects of compartmentalized sugar phosphate metabolism may differ in angiosperms and gymnosperms and furthermore underscore the contributions of endosymbiotic gene transfer and gene duplication to the nuclear complement of genes for enzymes of plant primary metabolism.
Similar content being viewed by others
References
Ansorge W, Sproat BS, Stegemann J, Schwager C: A non-radioactive automated method for DNA sequence determination. J Biochem Biophys Meth 13: 315–323 (1986).
Bartlett SG, Grossmann AR, Chua N-H: In vitro synthesis and uptake of cytoplasmically-synthesized chloroplast proteins. In: Edelmann M, Hallick RB, Chua N-H (eds) Methods in Chloroplast Molecular Biology, pp. 1081–1092. Elsevier Biomedical Press, Amsterdam (1982).
Brinkmann H, Martinez P, Quigley F, Martin W, Cerff R: Endosymbiotic origin and codon bias of the nuclear gene for chloroplast glyceraldehyde-3-phosphate dehydrogenase from maize. J Mol Evol 26: 24–33 (1987).
Brinkmann H, Cerff R, Salomon M, Soll J: Cloning and sequence analysis of cDNAs encoding the cytosolic precursors of subunits GapA and GapB of chloroplast glyceraldehyde-3-phosphate dehydrogenase from pea and spinach. Plant Mol Biol 13: 81–94 (1989).
Buchanan BB: Role of light in the regulation of chloroplast enzymes. Annu Rev Plant Physiol 31: 341–374 (1980).
Cerff R: Separation and purification of NAD- and NADP-linked glyceraldehyde-3-phosphate dehydrogenases from higher plants. In: Edelmann M, Hallick RB, Chua N-H (eds) Methods in Chloroplast Molecular Biology, pp. 683–694. Elsevier Biomedical Press, Amsterdam (1982).
Cerff R, Chambers S: Subunit structure of higher plant glyceraldehyde-3-phosphate dehydrogenases (EC 1.2.1.12 and 1.2.1.13). J Biol Chem 254: 6094–6098 (1979).
Cerff R, Kloppstech K: Structural diversity and differential light control of mRNAs coding for angiosperm glyceraldehyde-3-phosphate dehydrogenases. Proc Natl Acad Sci USA 79: 7624–7628 (1982).
Clermont S, Corbier C, Mely Y, Gerard D, Wonacott A, Branlant G: Determinants of coenzyme specificity in glyceraldehyde-3-phosphate dehydrogenase: role of the acidic residue in the fingerprint region of the nucleotide binding fold. Biochemistry 32: 10178–10184 (1993).
Corbier C, Clermont S, Billard P, Skarzynski T, Branlant C, Wonacott A, Branlant G: Probing the coenzyme specificity of glyceraldehyde-3-phosphate dehydrogenases by site directed mutagenisis. Biochemistry 29: 7101–7106 (1990).
Cséke C, Buchanan BB: Regulation of the formation and utilisation of photosynthate in leaves. Biochim Biophys Acta 853: 43–64 (1986).
Devereux J, Haeberli P, Smithies O: A comprehensive set of sequence analysis programms for the VAX. Nucl Acids Res 12: 387–395 (1984).
Dewdney J, Conley TR, Shih M-C, Goodmann H: Effects of blue and red light on expression of nuclear genes encoding chloroplast glyceraldehyde-3-phosphate dehydrogenase of Arabidopsis thaliana. Plant Physiol 103: 1115–1121 (1993).
Felsenstein J: Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39: 783–791 (1985).
Flügge U-I, Weber A, Fischer K, Lottspeich F, Eckerskorn C, Waegemann K, Soll J: The major chloroplast envelope polypeptide is the phosphate translocator and not the protein import receptor. Nature 353: 364–367 (1991).
Fothergill-Gilmore LA, Michels PAM: Evolution of glycolysis. Progr Biophys Mol Biol 59: 105–238 (1993).
Gottlieb LD: Conservation and duplication of isozymes in plants. Science 216: 373–380 (1982).
Harris JI, Waters M: Glyceraldehyde-3-phosphate dehydrogenase. In: Boyer PD (ed) The Enzymes, Vol. 13, pp. 1–50. Academic Press, New York (1976).
Heber U, Pon NG, Heber M: Localization of carboxydismutase and trisoephosphate dehydrogenases in the chloroplast. Plant Physiol 38: 355–360 (1963).
Jansson S, Meyer-Gauen G, Cerff R, Martin W: Nucleotide distribution in gymnosperm nuclear sequences suggest a model for GC-content change in land plant nuclear genomes. J Mol Evol 39: 34–46 (1994).
Kersanach R, Brinkmann H, Liaud M-F, Zhang D-X, Martin W, Cerff R: Five identical intron positions in acient duplicated genes of eubacterial origin. Nature 367: 387–389 (1994).
Kumada Y, Benson DR, Hillemann D, Hosted TJ, Rochefort DA, Thompson CJ, Wohlleben W, Tateno Y: Evolution of the glutamin synthase gene, one of the oldest existing and functioning genes. Proc Natl Acad Sci USA 90: 3009–3013 (1993).
Levy LM, Betts GF: The physiological significance of aggregation phenomena of NADP-dependent glyceraldehyde-3-phosphate dehydrogenase from spinach chloroplasts. Biochim Biophys Acta 832: 186–191 (1985).
Liaud M-F, Valentine C, Martin W, Bouget F-Y, Kloareg B, Cerff R: The evolutionary origin of red algae as deduced from the nuclear genes encoding cytosolic and chloroplast glyceraldehyde-3-phosphate dehydrogenases from Chondrus crispus. J Mol Evol 38: 319–327 (1994).
Liaud M-F, Zhang D-X, Cerff R: Differential intron loss and endosymbiotic transfer of chloroplast glyceraldehyde-3-phosphate dehydrogenase genes to the nucleus. Proc Natl Acad Sci USA 87: 8918–8922 (1990).
Li W-H, Wu C-I, Luo C-C: A new method for estimating synonymous and nonsynonymous rates of nucleotide substitution considering the relative likelihood of nucleotide and codon changes. Mol Biol Evol 2: 150–174 (1985).
Longstaff M, Raines CA, McMorrow E, Bradbeer JW, Dyer TA: Wheat phosphoglycerate kinase: evidence for recombination between the chloroplastic and cytosolic enzymes. Nucl Acids Res 17: 6569–6580 (1989).
Macdonald FD, Buchanan BB: The reductive pentose phosphate pathway and its regulation. In: Dennis DT, Turpin DH (eds) Plant Physiology, Biochemistry and Molecular Biology, pp. 249–262. Longman, Singapore (1990).
Martinez P, Martin W, Cerff R: Structure, evolution and anaerobic regulation of a nuclear gene encoding cytosolic glyceraldehyde-3-phosphate dehydrogenases from maize. J Mol Biol 208: 551–565 (1989).
Martin W, Cerff R: Prokaryotic features of a nucleus encoded enzyme: cDNA sequences for chloroplast and cytosolic glyceraldehyde-3-phosphate dehydrogenases from mustard (Sinapis alba). Eur J Biochem 159: 323–331 (1986).
Martin W, Brinkmann H, Savona C, Cerff R: Evidence for a chimaeric nature of nuclear genomes: eubacterial origin of eukaryotic glyceraldehyde-3-phosphate dehydrogenase genes. Proc Natl Acad Sci USA 90: 8692–8696 (1993).
Martin W, Gierl A, Saedler H: Molecular evidence for pre-Cretaceous angiosperm origins. Nature 339: 46–48 (1989).
Martin W, Lagrange T, Li Y-F, Bisanz-Seyer C, Mache R: Hypothesis for the evolutionary origin of the chloroplast ribosomal protein L21 of spinach. Curr Genet 18: 553–556 (1990).
Martin W, Lydiate D, Brinkmann H, Forkmann G, Saedler H, Cerff R: Molecular phylogenies in angiosperm evolution. Mol Biol Evol 10: 140–162 (1993).
Martin W, Nock S, Meyer-Gauen G, Häger K-P, Jensen U, Cerff R: A method for isolation of cDNA-quality mRNA from immature seeds of gymnosperm rich in polyphenolics. Plant Mol Biol 22: 555–556 (1993).
Martin W, Somerville CC, Loiseaux-de Goër S: Molecular phylogenies of plastid origins and algal evolution. J Mol Evol 35: 385–403 (1992).
Pelzer-Reith B, Penger A, Schnarrenberger C: Plant aldolase: cDNA and deduced amino-acid sequence of the chloroplast and cytosol enzyme from spinach. Plant Mol Biol 21: 331–340 (1993).
Peschek GA: Respiratory electron transport. In: Fay P, van Baalen C (eds) The Cyanobacteria, pp. 119–162. Elsevier, Amsterdam (1987).
Pupillo P, Faggiani R: Subunit structure of three glyceraldehyde-3-phosphate dehydrogenases of some flowering plants. Arch Biochem Biophys 194: 581–592 (1979).
Quigley F, Martin W, Cerff R: Intron conservation across the prokaryote-eukaryote boundry: Structure of the nuclear gene for chloroplast glyceraldehyde-3-phosphate dehydrogenase from maize. Proc Natl Acad Sci USA 85: 2672–2676 (1988).
Reith M, Munholland J: A high resolution gene map of the chloroplast genome of the red alga Porphyra purpurea. Plant Cell 5: 465–475 (1993).
Russell DA, Sachs MM: The maize cytosolic glyceraldehyde-3-phosphate dehydrogenase gene family: organ-specific expression and genetic analysis. Mol Gen Genet 229: 219–228 (1991).
Saitou N, Nei M: The neighbor-joining method: a new method for the reconstruction of phylogenetic trees. Mol Biol Evol 4: 406–425 (1987).
Sambrook J, Fritsch EF, Maniatis T: Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY (1989).
Scagliarini S, Trost P, Pupillo P, Valenti V: Light activation and molecular mass changes of NAD(P)-glyceraldehyde 3-phosphate dehydrogenase of spinach and maize leaves. Planta 190: 313–319 (1993).
Schnarrenberger C: Regulation and structure of isozymes of sugar phosphate metabolism in plants isozymes. In: Current Topics in Biological and Medical Research, vol. 16: Agriculture, Physiology, and Medicine, pp. 223–240. A.R. Liss, New York (1987).
Smith AJ: Modes of cyanobacterial carbon metabolism. In: Carr NG, Whitton BA (eds) The Biology of Cyanobacteria, pp. 47–85. Blackwell Scientific Publications, Oxford (1982).
Stitt M: The flux of carbon between the chloroplast and the cytosol. In: Dennis DT, Turpin DH (eds) Plant Physiology, Biochemistry and Molecular Biology, pp. 319–338. Longman, Singapore (1990).
Tingey S, Tsai F, Edwards J, Walker E, Coruzzi G: Chloroplast and cytosolic glutamine synthetase are encoded by homologous nuclear genes which are differentially expressed in vivo. J Biol Chem 263: 9651–9657 (1988).
Tsutsumi K, Kagaya Y, Hidaka S, Suzuki J, Tokairin J, Harai T, Hu D, Ishikawa K, Ejiri S: Structural analysis of the genes encoding chloroplastic and cytoplasmic aldolases in rice. Gene, in press.
Vermeglio A, Ravenel J, Peltier G: Chlororespiration: a respiratory activity in the thylakiod membrane of microalgae and higher plants. In: Weissner W, Robinson DG, Starr RC (eds) Experimental Phycology. 1. Cell Walls and Surfaces, Reproduction, Photosynthesis, pp. 188–205. Springer-Verlag, Berlin (1990).
von Heijne G, Stepphuhn J, Herrmann RG: Domain structure of mitochondrial and chloroplast targeting peptides. Eur J Biochem 180: 535–545 (1989).
Weeden NF: Genetic and biochemical implications of the endosymbiotic origin of the chloroplast. J Mol Evol 17: 133–139 (1981).
Yang Y, Kwon H-B, Peng H-P, Shih M-C: Stress responses and metabolic regulation of glyceraldehyde-3-phosphate dehydrogenase genes in Arabidopsis. Plant Physiol 101: 209–216 (1993).
Clausmeyer S, Klösgen R-B, Herrmann RG: Protein import into chloroplasts: The hydrophilic lumenal proteins exhibit unexpected import and sorting specificities in spite of structurally conserved transit peptides. J Biol Chem 268: 13869–13876 (1993).
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Meyer-Gauen, G., Schnarrenberger, C., Cerff, R. et al. Molecular characterization of a novel, nuclear-encoded, NAD+-dependent glyceraldehyde-3-phosphate dehydrogenase in plastids of the gymnosperm Pinus sylvestris L.. Plant Mol Biol 26, 1155–1166 (1994). https://doi.org/10.1007/BF00040696
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/BF00040696